
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,546,548 – 6,546,641 |
| Length | 93 |
| Max. P | 0.849176 |

| Location | 6,546,548 – 6,546,641 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -24.13 |
| Energy contribution | -25.60 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
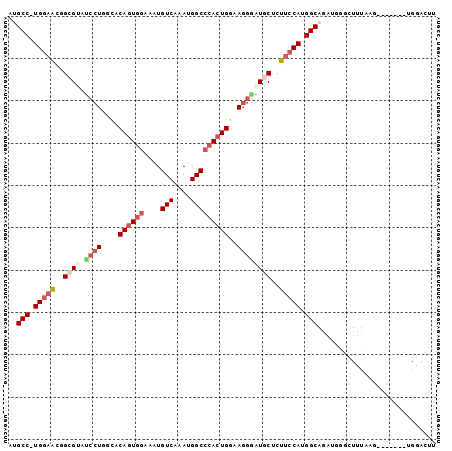
>2L_DroMel_CAF1 6546548 93 + 22407834 AUGCC-UGGAACGGCGUCUCCUGGCACAGUGGAAAUGUCAAAUGGCCCACUGGAAGGGAUGCUCUUCCAUGGCAGAUGGGCUUUAAG-------UGGACUU .((((-(((((.((((((.(((....((((((....(((....)))))))))..))))))))).))))).)))).............-------....... ( -37.20) >DroSec_CAF1 18497 93 + 1 AUGCC-UGGAACGGCGUGUCCUGGCACAGUGGAAAUGUCAAAUGGCCCACUGGAAGGUAUGCUCUUCCAUGGCAGAUGUGCUUUAAG-------UGGACUU .((((-(((((.((((((.(((....((((((....(((....)))))))))..))))))))).))))).))))...((.(......-------.).)).. ( -35.50) >DroSim_CAF1 20436 93 + 1 AUGCC-UGGAACGGCGUGUCCUGGCACAGUGGAAAUGUCAAAUGGCCCACUGGAAGGAAUGCUCUUCCAUGGCAGAUGGGCUUUAUG-------UGGACUU .((((-(((((.(((((.((((....((((((....(((....)))))))))..))))))))).))))).)))).............-------....... ( -34.80) >DroEre_CAF1 19107 92 + 1 AAGCC-UGGAACUGCGGAUCCUGGCACAGUGGAAAUGUCAAAGGGCCCACUGGAAGGGAUGCCUUUCCAUGGCAGAUACGCUUUGAG-------UGGACU- ..(((-(((((..(.(.(((((..(.((((((....(((....))))))))))..))))).)).))))).))).....(((.....)-------))....- ( -32.50) >DroYak_CAF1 19601 93 + 1 AUGCC-UGGAACUGCGGAUCCUGGCACAGUGGAAAUGUCAAAGGGCCCACUGAAAGGGAUACUCUUCCAUGGCAAAUGGGCUUUAAG-------UGGACUU .((((-(((((..(...(((((....((((((....(((....)))))))))...))))).)..))))).)))).............-------....... ( -27.80) >DroAna_CAF1 20439 100 + 1 GGGCCGUGCAGCAG-GAUCCCUGGUGCAAUGGGAAUGUCAAAUGGCGAACUGGAAUAGUUUCGUUCUCAUGGCAGCUGCCUUUGGAGCUAGUUGGGGAUUU ..(((.((((.(((-(...)))).))))(((((((((((....)))((((((...)))))).)))))))))))((((.((...))))))............ ( -31.70) >consensus AUGCC_UGGAACGGCGUAUCCUGGCACAGUGGAAAUGUCAAAUGGCCCACUGGAAGGGAUGCUCUUCCAUGGCAGAUGGGCUUUAAG_______UGGACUU ..(((.(((((..((((.((((....((((((....(((....)))))))))..))))))))..))))).)))............................ (-24.13 = -25.60 + 1.47)


| Location | 6,546,548 – 6,546,641 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.15 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
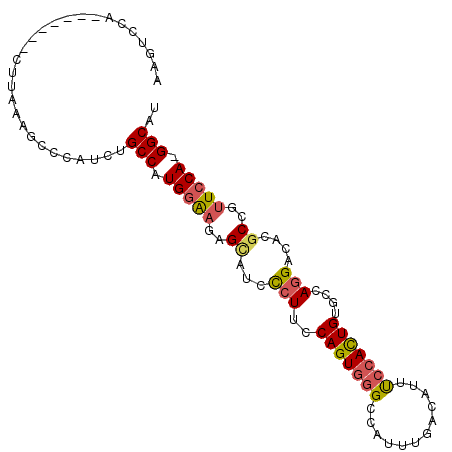
>2L_DroMel_CAF1 6546548 93 - 22407834 AAGUCCA-------CUUAAAGCCCAUCUGCCAUGGAAGAGCAUCCCUUCCAGUGGGCCAUUUGACAUUUCCACUGUGCCAGGAGACGCCGUUCCA-GGCAU .......-------.............((((.(((((..((.(((((.((((((((............))))))).)..))).)).))..)))))-)))). ( -27.70) >DroSec_CAF1 18497 93 - 1 AAGUCCA-------CUUAAAGCACAUCUGCCAUGGAAGAGCAUACCUUCCAGUGGGCCAUUUGACAUUUCCACUGUGCCAGGACACGCCGUUCCA-GGCAU ..((((.-------......(((((.......((((((.......))))))(((((............))))))))))..))))..(((......-))).. ( -26.80) >DroSim_CAF1 20436 93 - 1 AAGUCCA-------CAUAAAGCCCAUCUGCCAUGGAAGAGCAUUCCUUCCAGUGGGCCAUUUGACAUUUCCACUGUGCCAGGACACGCCGUUCCA-GGCAU ..((((.-------......(((((.(((....((((.....))))...))))))))....((.(((.......))).))))))..(((......-))).. ( -29.00) >DroEre_CAF1 19107 92 - 1 -AGUCCA-------CUCAAAGCGUAUCUGCCAUGGAAAGGCAUCCCUUCCAGUGGGCCCUUUGACAUUUCCACUGUGCCAGGAUCCGCAGUUCCA-GGCUU -......-------..............(((.(((((..((((((...((((((((............))))))).)...))))..))..)))))-))).. ( -26.30) >DroYak_CAF1 19601 93 - 1 AAGUCCA-------CUUAAAGCCCAUUUGCCAUGGAAGAGUAUCCCUUUCAGUGGGCCCUUUGACAUUUCCACUGUGCCAGGAUCCGCAGUUCCA-GGCAU .......-------.............((((.(((((..((((((....(((((((............))))))).....))))..))..)))))-)))). ( -24.90) >DroAna_CAF1 20439 100 - 1 AAAUCCCCAACUAGCUCCAAAGGCAGCUGCCAUGAGAACGAAACUAUUCCAGUUCGCCAUUUGACAUUCCCAUUGCACCAGGGAUC-CUGCUGCACGGCCC ............((((((...)).))))((((((.((((............))))..))).............((((.((((...)-))).)))).))).. ( -21.10) >consensus AAGUCCA_______CUUAAAGCCCAUCUGCCAUGGAAGAGCAUCCCUUCCAGUGGGCCAUUUGACAUUUCCACUGUGCCAGGACACGCCGUUCCA_GGCAU ............................(((.(((((..((...(((..(((((((............)))))))....)))....))..))))).))).. (-18.81 = -19.15 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:11 2006