
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,540,931 – 6,541,074 |
| Length | 143 |
| Max. P | 0.935705 |

| Location | 6,540,931 – 6,541,046 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6540931 115 + 22407834 ACACACAC-GCUUACGCACAAUUAUUAACACUUAUUAAUGUUGAGGUUUCCAUAUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCGAGUGCCAAACAGCUGCCUCAAGUGC---- ........-......((((...((((((......))))))(((((((...........(((((((....(((((......))))).....).)))))).......)))))))))))---- ( -26.37) >DroVir_CAF1 97613 106 + 1 ----------CACACACACAAUUAUUAACACUUAUUAAUGUUGAGGUUUUUAUAUAGAUGGCACCCCAGGGCCUCAAUUAAAGUC--CAAAAGUGCCA-ACAGCUGCCUCAAGUGGCU-U ----------......(((...((((((......))))))(((((((...........((((((....((((..........)))--)....))))))-......))))))))))...-. ( -23.73) >DroPse_CAF1 13643 103 + 1 -------------AUACACAAUUAUUAACACUUAUUAAUGUUGAGGUUUUUGCGUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCAAGUGCCAAACAGCUGCCUCAAGUGC---- -------------...(((...((((((......))))))(((((((....((.....((((((.....(((((......))))).......))))))....)).)))))))))).---- ( -27.40) >DroGri_CAF1 18036 101 + 1 ----------C---UACACAAUUAUUAACACUUAUUAAUGUUGAGGUUUUUA--UAGAUGGCACCCAGGGGCUUCAAUUAAAGCC--CA-AAGUGCCA-ACAGCUGCCUCAAGCGCCAGU ----------.---..........((((((........))))))(((.....--..(.((((((....((((((......)))))--).-..))))))-.).(((......))))))... ( -29.10) >DroMoj_CAF1 123408 107 + 1 UCA-ACACUCCACACACACAAUUAUUAACACUUAUUAUUGUUGAGGUUUUUAUAUAGAUGGCACCCCUGGGCCUCAAUUAAAUUC--CA--AGUGCCA-ACAGCUGCUUCAA-------U ...-...................................((((((((...........((((((...((((............))--))--.))))))-......)))))))-------) ( -15.53) >DroAna_CAF1 13945 110 + 1 A-----GC-AUAUACACACAAUUAUUAACACUUAUUAAUGUUGAGGUUUUCGUAUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCGAGUGCCAAACAGCUGGCUCAAGUGC---- .-----.(-(((((.((.......((((((........)))))).......)).))))))((((.....(((((......))))).......(.((((......)))).)..))))---- ( -22.44) >consensus __________CA_ACACACAAUUAUUAACACUUAUUAAUGUUGAGGUUUUUAUAUAGAUGGCACCCCAGGGCUUAAAUUAAAGUC__CACAAGUGCCA_ACAGCUGCCUCAAGUGC____ ........................................(((((((...........((((((.....(((((......))))).......)))))).......)))))))........ (-17.31 = -17.70 + 0.39)

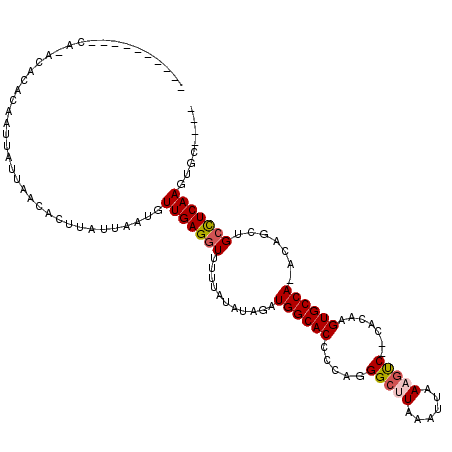

| Location | 6,540,931 – 6,541,046 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -18.03 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6540931 115 - 22407834 ----GCACUUGAGGCAGCUGUUUGGCACUCGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUAUAUGGAAACCUCAACAUUAAUAAGUGUUAAUAAUUGUGCGUAAGC-GUGUGUGU ----((((..((((...((((.((((((((.((..(.(((......)))).)).)))))))).....))))...))))((((((....)))))).........((....))-)))).... ( -33.20) >DroVir_CAF1 97613 106 - 1 A-AGCCACUUGAGGCAGCUGU-UGGCACUUUUG--GACUUUAAUUGAGGCCCUGGGGUGCCAUCUAUAUAAAAACCUCAACAUUAAUAAGUGUUAAUAAUUGUGUGUGUG---------- .-.(((((..((((....(((-((((((((..(--(.((((....)))).))..)))))))).....)))....))))((((((....)))))).......))).))...---------- ( -27.20) >DroPse_CAF1 13643 103 - 1 ----GCACUUGAGGCAGCUGUUUGGCACUUGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUACGCAAAAACCUCAACAUUAAUAAGUGUUAAUAAUUGUGUAU------------- ----((((..((((..(((((.((((((((.((..(.(((......)))).)).)))))))).))).)).....))))((((((....)))))).......))))..------------- ( -28.60) >DroGri_CAF1 18036 101 - 1 ACUGGCGCUUGAGGCAGCUGU-UGGCACUU-UG--GGCUUUAAUUGAAGCCCCUGGGUGCCAUCUA--UAAAAACCUCAACAUUAAUAAGUGUUAAUAAUUGUGUA---G---------- ....((((..((((.......-((((((((-.(--((((((....)))))))..))))))))....--......))))((((((....)))))).......)))).---.---------- ( -33.93) >DroMoj_CAF1 123408 107 - 1 A-------UUGAAGCAGCUGU-UGGCACU--UG--GAAUUUAAUUGAGGCCCAGGGGUGCCAUCUAUAUAAAAACCUCAACAAUAAUAAGUGUUAAUAAUUGUGUGUGUGGAGUGU-UGA .-------......((((...-(((((((--((--(..(((....)))..)))..)))))))((((((((.........(((((.((........)).))))).))))))))..))-)). ( -23.20) >DroAna_CAF1 13945 110 - 1 ----GCACUUGAGCCAGCUGUUUGGCACUCGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUAUACGAAAACCUCAACAUUAAUAAGUGUUAAUAAUUGUGUGUAUAU-GC-----U ----((((....(((((....))))).((((((..(.(((......))))))))))))))((((((((((........((((((....))))))........)))))))))-).-----. ( -30.79) >consensus ____GCACUUGAGGCAGCUGU_UGGCACUUGGG__GACUUUAAUUGAAGCCCUGGGGUGCCAUAUAUAUAAAAACCUCAACAUUAAUAAGUGUUAAUAAUUGUGUGU_UG__________ ....((((..((((....(((.((((((((.((....(((......)))..)).)))))))).....)))....))))((((((....)))))).......))))............... (-18.03 = -18.90 + 0.87)


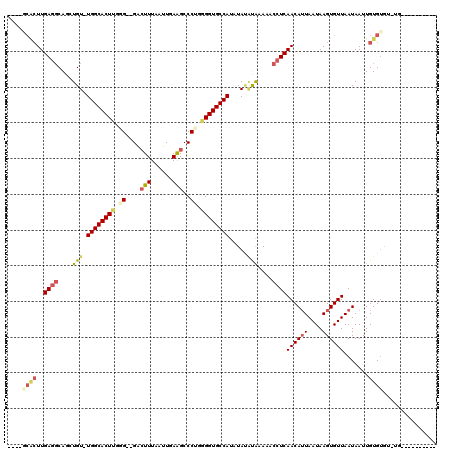
| Location | 6,540,970 – 6,541,074 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
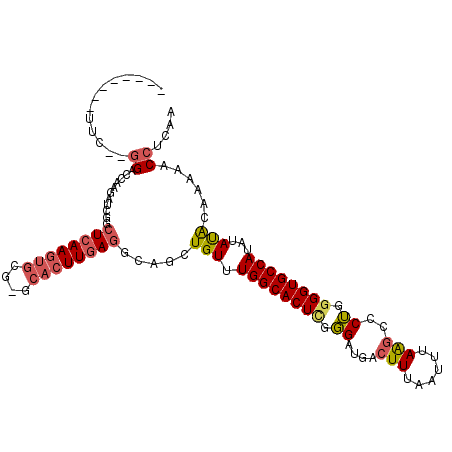
>2L_DroMel_CAF1 6540970 104 + 22407834 UUGAGGUUUCCAUAUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCGAGUGCCAAACAGCUGCCUCAAGUGC-CGCACUUGAGCCGAUCUGGAUCCCGGAA-------- ....((..((((......(((((((....(((((......))))).....).))))))......((.((((((((.-..)))))))).))...))))..))....-------- ( -31.90) >DroPse_CAF1 13670 100 + 1 UUGAGGUUUUUGCGUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCAAGUGCCAAACAGCUGCCUCAAGUGC-CGCACUUGAG----UCUCGCUCCUCGAC-------- ((((((.....(((....((((((.....(((((......))))).......)))))).........((((((((.-..))))))))----...))).)))))).-------- ( -32.10) >DroYak_CAF1 13585 111 + 1 UUGAGGUUUCGAUAUAGAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCGAGUGCCAAACAGCUGCCUCAAGUGU-CGCACUUGAGCCGAUCUGGGUCC-GGAAUCCGGAAU (((.(((((((((.(((((((((((....(((((......))))).....).))))).......((.((((((((.-..)))))))).))))))).)).)-)))))))))... ( -33.70) >DroMoj_CAF1 123447 91 + 1 UUGAGGUUUUUAUAUAGAUGGCACCCCUGGGCCUCAAUUAAAUUC--CA--AGUGCCA-ACAGCUGCUUCAA----UCGCACUUCAGCCCCUGUUGGCCC------------- ....((..((((....((.(((.((...)))))))...))))..)--).--...((((-((((..(((....----.........)))..))))))))..------------- ( -24.62) >DroAna_CAF1 13979 98 + 1 UUGAGGUUUUCGUAUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCGAGUGCCAAACAGCUGGCUCAAGUGC-GGCACUUGAGCCGAUCAUCAUC-------------- ....(((..((.......(((((((....(((((......))))).....).)))))).......((((((((((.-..))))))))))))..)))...-------------- ( -33.00) >DroPer_CAF1 13701 100 + 1 UUGAGGUUUUUGCGUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCAAGUGCCAAACAGCUGCCUCAAGUGC-CGCACUUGAG----UGUCGCUCCUCGAA-------- ((((((............((((((.....(((((......))))).......))))))...(((.((((((((((.-..))))))))----.)).))))))))).-------- ( -34.60) >consensus UUGAGGUUUUUAUAUAUAUGGCACCCCAGGGCUUAAAUUAAAGUCAUCCCGAGUGCCAAACAGCUGCCUCAAGUGC_CGCACUUGAGCCGAUCUCGAUCC__GAA________ ..................((((((.....(((((......))))).......)))))).........((((((((....)))))))).......................... (-19.70 = -20.57 + 0.86)



| Location | 6,540,970 – 6,541,074 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6540970 104 - 22407834 --------UUCCGGGAUCCAGAUCGGCUCAAGUGCG-GCACUUGAGGCAGCUGUUUGGCACUCGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUAUAUGGAAACCUCAA --------....((..((((....(.((((((((..-.)))))))).)...(((.((((((((.((..(.(((......)))).)).)))))))).)))..))))..)).... ( -37.50) >DroPse_CAF1 13670 100 - 1 --------GUCGAGGAGCGAGA----CUCAAGUGCG-GCACUUGAGGCAGCUGUUUGGCACUUGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUACGCAAAAACCUCAA --------...(((((((....----((((((((..-.))))))))...)))((.((((((((.((..(.(((......)))).)).))))))))...)).......)))).. ( -35.80) >DroYak_CAF1 13585 111 - 1 AUUCCGGAUUCC-GGACCCAGAUCGGCUCAAGUGCG-ACACUUGAGGCAGCUGUUUGGCACUCGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUCUAUAUCGAAACCUCAA ..(((((...))-)))..(((...(.((((((((..-.)))))))).)..)))..((((((((.((..(.(((......)))).)).)))))))).................. ( -36.00) >DroMoj_CAF1 123447 91 - 1 -------------GGGCCAACAGGGGCUGAAGUGCGA----UUGAAGCAGCUGU-UGGCACU--UG--GAAUUUAAUUGAGGCCCAGGGGUGCCAUCUAUAUAAAAACCUCAA -------------(((((..((((.((..((((((..----.....))).)).)-..)).))--))--...((.....))))))).(((((...............))))).. ( -27.36) >DroAna_CAF1 13979 98 - 1 --------------GAUGAUGAUCGGCUCAAGUGCC-GCACUUGAGCCAGCUGUUUGGCACUCGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUAUACGAAAACCUCAA --------------..(((.(...((((((((((..-.))))))))))...(((.((((((((.((..(.(((......)))).)).)))))))).)))........).))). ( -34.60) >DroPer_CAF1 13701 100 - 1 --------UUCGAGGAGCGACA----CUCAAGUGCG-GCACUUGAGGCAGCUGUUUGGCACUUGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUACGCAAAAACCUCAA --------...(((((((..(.----((((((((..-.)))))))))..)))((.((((((((.((..(.(((......)))).)).))))))))...)).......)))).. ( -36.20) >consensus ________UUC__GGACCAAGAUCGGCUCAAGUGCG_GCACUUGAGGCAGCUGUUUGGCACUCGGGAUGACUUUAAUUUAAGCCCUGGGGUGCCAUAUAUACAAAAACCUCAA .............((...........((((((((....)))))))).....(((.((((((((.((....(((......)))..)).))))))))...)))......)).... (-22.94 = -23.25 + 0.31)

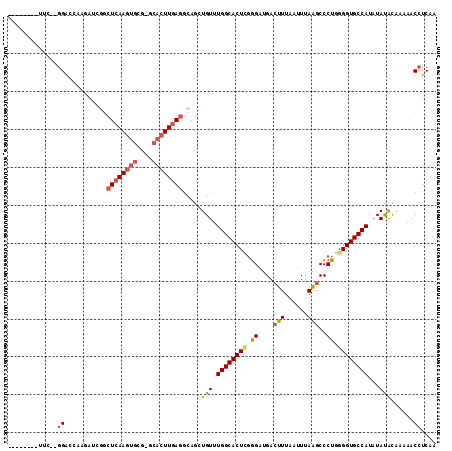
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:09 2006