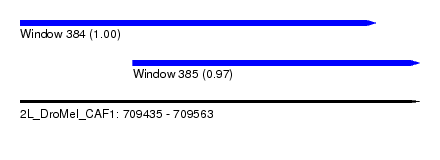
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 709,435 – 709,563 |
| Length | 128 |
| Max. P | 0.997895 |

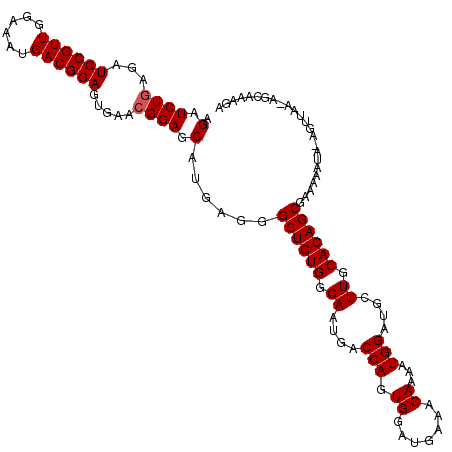
| Location | 709,435 – 709,549 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 709435 114 + 22407834 AGAUCUAAGAUCCGUUGGAAAUGACGGAGUGAACGGAGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAUGCUUGCACAGCGAAAAAUA-AGUUAA-AGCAAAGA ...(((....((((((......))))))((.(((...........((((((.((..(.(((.((.......))...))).)..)).))))))........-.)))..-.))..))) ( -24.25) >DroSec_CAF1 88465 114 + 1 AGAUCUGAGAUCCGUUGGAAAUGACGGAGCGAACGGAGCAUGAGGGCUGUGGGAAUGACCAGUGGAUAAAACAAAAUGGAUGCUUGCACAGCGAAAAAUA-AGUUAA-AACAAAGA .(.((((.(.((((((......)))))).)...)))).)......((((((.((..(.(((.((.......))...))).)..)).))))))........-......-........ ( -26.70) >DroSim_CAF1 93460 114 + 1 AGAUCUGAGAUCCGUUGGAGAUGACGGAGCGAACGGAGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAUUCUUGCACAGCGAAAAAUA-AGUUAA-AGCAAAGA .(.((((.(.((((((......)))))).)...)))).)......(((((((((((..(((.((.......))...))))))))..))))))........-......-........ ( -26.50) >DroEre_CAF1 97828 115 + 1 AGAUCUGAGAUCCGUUGGAAAUGACGGAGUGAACGGAGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAAGCUUGCACAGCGAAAAAAAGAGUUUA-AGCAAAGA .(.((((...((((((......)))))).....)))).)......((((((.((.(..(((.((.......))...)))..).)).))))))...............-........ ( -25.90) >DroYak_CAF1 93933 114 + 1 AGAUCUGAGAUCCGUUCGAAAUGACGGAGUGGACGGAGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAAGCUUGCACAGCGAAAAAAAG--UUACUGCCAAAGA ...(((....((((((......)))))).(((.((((((......((((((.((.(..(((.((.......))...)))..).)).))))))........)--)).)))))).))) ( -27.34) >consensus AGAUCUGAGAUCCGUUGGAAAUGACGGAGUGAACGGAGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAUGCUUGCACAGCGAAAAAUA_AGUUAA_AGCAAAGA .(.((((...((((((......)))))).....)))).)......((((((.((....(((.((.......))...)))....)).))))))........................ (-24.90 = -25.10 + 0.20)

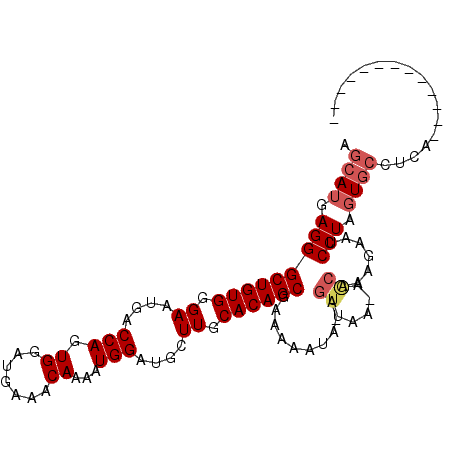
| Location | 709,471 – 709,563 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 709471 92 + 22407834 AGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAUGCUUGCACAGCGAAAAAUA-AGUUAA-AGCAAAGAAUCCUAGUGCCUCA------------- .((((.(((((((((.((..(.(((.((.......))...))).)..)).))))))........-.((...-.)).......))).))))....------------- ( -23.60) >DroSec_CAF1 88501 90 + 1 AGCAUGAGGGCUGUGGGAAUGACCAGUGGAUAAAACAAAAUGGAUGCUUGCACAGCGAAAAAUA-AGUUAA-AACAAAGAAUCCUAGUGCCU--------------- .((((.(((((((((.((..(.(((.((.......))...))).)..)).))))))........-.((...-.)).......))).))))..--------------- ( -23.00) >DroSim_CAF1 93496 92 + 1 AGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAUUCUUGCACAGCGAAAAAUA-AGUUAA-AGCAAAGAAUCCUAGUGCCUCA------------- .((((.((((((((((((((..(((.((.......))...))))))))..))))))........-.((...-.)).......))).))))....------------- ( -23.40) >DroEre_CAF1 97864 106 + 1 AGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAAGCUUGCACAGCGAAAAAAAGAGUUUA-AGCAAAGAAUCCUACUGCCCCAAUGCCUCAAGGCA ....((.((((.((((((....(((.((.......))...)))..(((((.((.............)).))-)))......)))))).)))))).((((....)))) ( -31.72) >DroYak_CAF1 93969 86 + 1 AGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAAGCUUGCACAGCGAAAAAAAG--UUACUGCCAAAGAAUCCUAGU------------------- ......(((((((((.((.(..(((.((.......))...)))..).)).)))))).........--...((.....))...)))...------------------- ( -17.50) >consensus AGCAUGAGGGCUGUGGGAAUGACCAGUGGAUGAAACAAAAUGGAUGCUUGCACAGCGAAAAAUA_AGUUAA_AGCAAAGAAUCCUAGUGCCUCA_____________ .((((.(((((((((.((....(((.((.......))...)))....)).))))))..........((.....)).......))).))))................. (-19.02 = -19.90 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:57 2006