
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
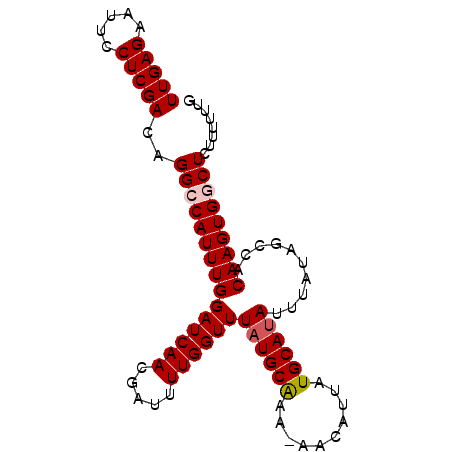
| Location | 6,523,538 – 6,523,630 |
| Length | 92 |
| Max. P | 0.970540 |

| Location | 6,523,538 – 6,523,630 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6523538 92 + 22407834 UUGAGAAUUCCUCGACAGGCCAUUUGGAUCAACGAUUUUGGUUUAUGCAAAAAACUUUAUGCACAUUUAUAGCCACAAGUGCCUCUUUUUUG (((((.....))))).((((.(((((((((...)))).(((((..((((.((....)).)))).......))))))))))))))........ ( -16.00) >DroSec_CAF1 10617 91 + 1 UUGAGAAUUCCUCGACAGGCCAUUUGGAUCAACGAUUUUGGUUUAUGCAAA-AACAUUAUGCAUAUUUAUAGCCACAAGUGGCUCUUUUUUU (((((.....)))))..(((((((((((((...)))).(((((((((((..-.......)))))).....))))))))))))))........ ( -22.80) >DroSim_CAF1 10522 91 + 1 UUGAGAAUUCCUCGACAGGCCAUUUGGAUCAACGAUUUUGGUUUAUGCAAA-AACAUUAUGCAUAUUUAUAGCCACAAGUGGCUCUUUUUUG (((((.....)))))..(((((((((((((...)))).(((((((((((..-.......)))))).....))))))))))))))........ ( -22.80) >DroEre_CAF1 8079 91 + 1 UUGAGAAUUCCUCGACAGGCCAUUUGGAUCAACAAUUUUGGUUUAUGCGAA-AACACUAUGCAUAUUUAUAGCCGCAAGUGGCUCUUUUUUG (((((.....)))))..(((((((((((((((.....))))))((((((..-.......))))))..........)))))))))........ ( -20.60) >DroYak_CAF1 9762 91 + 1 UUGAGAAUUCCUCGACAGGCCAUUUGGAUCAACGAUUUUGGUUUAUGCAAA-AACAUUAUGCAUAUUUAUAGCCACAAGUGUCUCUUUUUUG (((((.....))))).((((.(((((((((...)))).(((((((((((..-.......)))))).....))))))))))))))........ ( -16.30) >consensus UUGAGAAUUCCUCGACAGGCCAUUUGGAUCAACGAUUUUGGUUUAUGCAAA_AACAUUAUGCAUAUUUAUAGCCACAAGUGGCUCUUUUUUG (((((.....)))))..(((((((((((((((.....))))))((((((..........))))))..........)))))))))........ (-18.10 = -18.54 + 0.44)


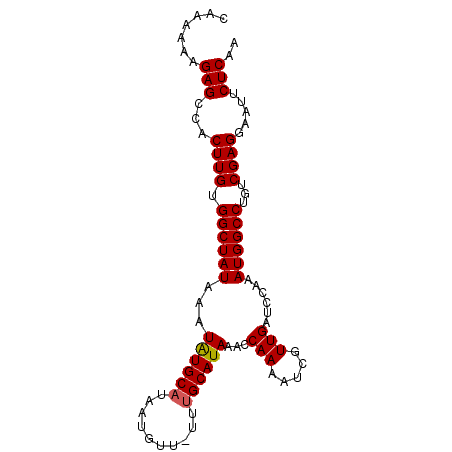
| Location | 6,523,538 – 6,523,630 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -17.84 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6523538 92 - 22407834 CAAAAAAGAGGCACUUGUGGCUAUAAAUGUGCAUAAAGUUUUUUGCAUAAACCAAAAUCGUUGAUCCAAAUGGCCUGUCGAGGAAUUCUCAA .......((((..((((.((((((...((((((..........))))))...(((.....)))......))))))...))))....)))).. ( -19.90) >DroSec_CAF1 10617 91 - 1 AAAAAAAGAGCCACUUGUGGCUAUAAAUAUGCAUAAUGUU-UUUGCAUAAACCAAAAUCGUUGAUCCAAAUGGCCUGUCGAGGAAUUCUCAA ........(((((....))))).....((((((.......-..))))))........((.(((((((....))...))))).))........ ( -19.00) >DroSim_CAF1 10522 91 - 1 CAAAAAAGAGCCACUUGUGGCUAUAAAUAUGCAUAAUGUU-UUUGCAUAAACCAAAAUCGUUGAUCCAAAUGGCCUGUCGAGGAAUUCUCAA ........(((((....))))).....((((((.......-..))))))........((.(((((((....))...))))).))........ ( -19.00) >DroEre_CAF1 8079 91 - 1 CAAAAAAGAGCCACUUGCGGCUAUAAAUAUGCAUAGUGUU-UUCGCAUAAACCAAAAUUGUUGAUCCAAAUGGCCUGUCGAGGAAUUCUCAA .......(((...(((((((((((...(((((........-...)))))........(((......))))))))).).)))).....))).. ( -16.60) >DroYak_CAF1 9762 91 - 1 CAAAAAAGAGACACUUGUGGCUAUAAAUAUGCAUAAUGUU-UUUGCAUAAACCAAAAUCGUUGAUCCAAAUGGCCUGUCGAGGAAUUCUCAA .......((((..((((.((((((...((((((.......-..))))))...(((.....)))......))))))...))))....)))).. ( -20.90) >consensus CAAAAAAGAGCCACUUGUGGCUAUAAAUAUGCAUAAUGUU_UUUGCAUAAACCAAAAUCGUUGAUCCAAAUGGCCUGUCGAGGAAUUCUCAA .......(((...((((.((((((...((((((..........))))))...(((.....)))......))))))...)))).....))).. (-17.84 = -17.88 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:00 2006