| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 700,584 – 700,675 |
| Length | 91 |
| Max. P | 0.803218 |

| Location | 700,584 – 700,675 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
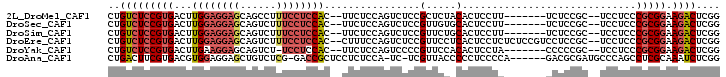
>2L_DroMel_CAF1 700584 91 + 22407834 CUGUCUCCGUGACUUGGAGGAGCAGCCUUUCCUCCAC--UUCUCCAGUCUCCGCUCUACACUCCUU-------UCUCCGC--UCCUCCCGCGGAAGACUCGG ..(((.....))).((((((((......)))))))).--....(((((((((((............-------.......--.......)))).))))).)) ( -25.47) >DroSec_CAF1 79694 91 + 1 CUGUCUCCGUGACUUGGAGGAGCAGUCUUUCCUCCAC--UUCUCCAGUCUCCGUUGUGCACUCCUU-------UCUCCGC--UCCUCCCGCGGAAGACUCGG ..(((((((((....((((((((((.........(((--................)))........-------.))..))--))))))))))).)))).... ( -27.16) >DroSim_CAF1 84663 91 + 1 CUGUCUCCGUGACUUGGAGGAGCAGUCUUUCCUCCAC--UUCUCCAGUCUCCGUUCUGCACUCCUU-------UCUCCGC--UCCUCCCGCGGAAGACUCGG ..(((((((((....(((((((((((.........))--)....(((........)))........-------.....))--))))))))))).)))).... ( -27.60) >DroEre_CAF1 88954 98 + 1 CUGUCUCCGUGACUUGGAGGAGCAGUCUUUCCUCCAC--CUUUCCAGUCUCCGUUCCUCACUCCUCUCUCCGUCCUCCGC--UCCUCCCGCGGAAGACUCGG ......(((.((((((((((((......)))))))).--......))))......................(((.(((((--.......))))).))).))) ( -27.21) >DroYak_CAF1 85050 90 + 1 CUGUCUCCGUGACUUGAAGGAGCAGUCU-UCCUCCAC--UUCUCCAGUCCCCGUUCCACACUCCUA-------CCCCCGC--UCCUCCCGCGGAAGACUCGG ..(((((((((....(.(((((((((..-......))--)............(.....).......-------.....))--)))).)))))).)))).... ( -19.10) >DroAna_CAF1 44559 93 + 1 CUGACUUCGUGACGUGGAGGAGCUGUCUCG-GACCGCUCCUCUCCA-UC-UCGUUACCCCCUCCCCA------GACGCGAUGCCCAGCCUCGCAAAUCUCGG ........((((((.((((((((.(((...-))).))))))))...-..-.))))))......((.(------((.((((.((...)).))))...))).)) ( -30.20) >consensus CUGUCUCCGUGACUUGGAGGAGCAGUCUUUCCUCCAC__UUCUCCAGUCUCCGUUCCACACUCCUU_______UCUCCGC__UCCUCCCGCGGAAGACUCGG ..(((((((((...((((((((......))))))))................(.....).............................))))).)))).... (-15.76 = -16.73 + 0.98)
| Location | 700,584 – 700,675 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -21.15 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
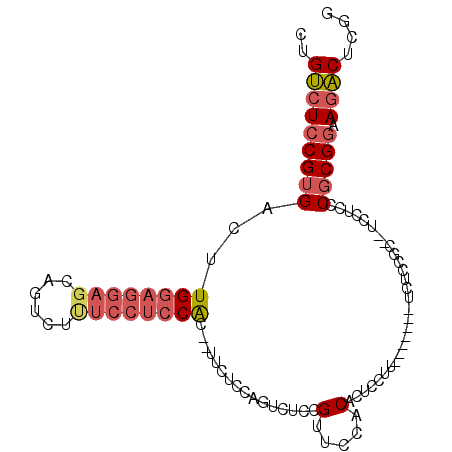
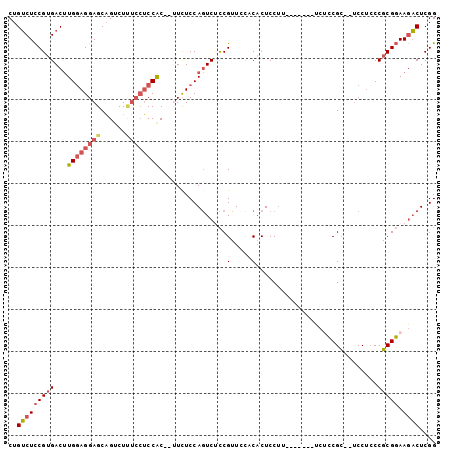
>2L_DroMel_CAF1 700584 91 - 22407834 CCGAGUCUUCCGCGGGAGGA--GCGGAGA-------AAGGAGUGUAGAGCGGAGACUGGAGAA--GUGGAGGAAAGGCUGCUCCUCCAAGUCACGGAGACAG ((.(((((.((((...(.(.--.(.....-------...)..).)...))))))))))).((.--.(((((((........)))))))..))..(....).. ( -31.90) >DroSec_CAF1 79694 91 - 1 CCGAGUCUUCCGCGGGAGGA--GCGGAGA-------AAGGAGUGCACAACGGAGACUGGAGAA--GUGGAGGAAAGACUGCUCCUCCAAGUCACGGAGACAG ((.((((((((((.......--)))))..-------.....((.....))..))))))).((.--.(((((((........)))))))..))..(....).. ( -31.40) >DroSim_CAF1 84663 91 - 1 CCGAGUCUUCCGCGGGAGGA--GCGGAGA-------AAGGAGUGCAGAACGGAGACUGGAGAA--GUGGAGGAAAGACUGCUCCUCCAAGUCACGGAGACAG ((.((((((((((.......--)))))..-------.....((.....))..))))))).((.--.(((((((........)))))))..))..(....).. ( -31.40) >DroEre_CAF1 88954 98 - 1 CCGAGUCUUCCGCGGGAGGA--GCGGAGGACGGAGAGAGGAGUGAGGAACGGAGACUGGAAAG--GUGGAGGAAAGACUGCUCCUCCAAGUCACGGAGACAG (((.(((((((((.......--)))))))))((((.(((.(((......(.....((....))--.....).....))).)))))))......)))...... ( -36.30) >DroYak_CAF1 85050 90 - 1 CCGAGUCUUCCGCGGGAGGA--GCGGGGG-------UAGGAGUGUGGAACGGGGACUGGAGAA--GUGGAGGA-AGACUGCUCCUUCAAGUCACGGAGACAG ....((((.(((.(((((((--((((...-------.....((.....))(....).......--........-...))))))))))....).))))))).. ( -27.70) >DroAna_CAF1 44559 93 - 1 CCGAGAUUUGCGAGGCUGGGCAUCGCGUC------UGGGGAGGGGGUAACGA-GA-UGGAGAGGAGCGGUC-CGAGACAGCUCCUCCACGUCACGAAGUCAG ((.((((..((((.((...)).)))))))------).))..........((.-((-((..(((((((.(((-...))).)))))))..)))).))....... ( -36.70) >consensus CCGAGUCUUCCGCGGGAGGA__GCGGAGA_______AAGGAGUGCAGAACGGAGACUGGAGAA__GUGGAGGAAAGACUGCUCCUCCAAGUCACGGAGACAG ....(((((((((.........)))).......................((..((((((((....((((........))))..))))).))).))))))).. (-21.15 = -20.27 + -0.88)

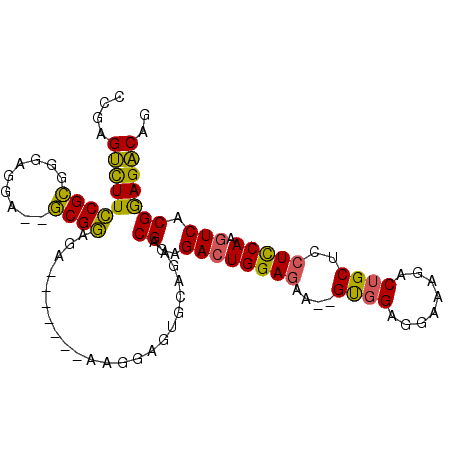
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:55 2006