
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
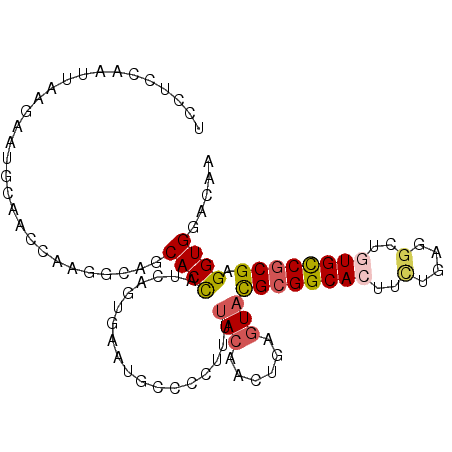
| Location | 6,485,227 – 6,485,326 |
| Length | 99 |
| Max. P | 0.565338 |

| Location | 6,485,227 – 6,485,326 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6485227 99 + 22407834 UCCUCCAAUUAAGAAUGCAGCCAAGGCAGCACCAUCAGUGAAUGCCCCUUUUACAACUGAGUACGCGGCACUUCUUCGUCUUUGCCGCGAGGUGGACAA (((.((.........(((.((....)).)))...(((((.((........))...)))))...(((((((............))))))).)).)))... ( -27.60) >DroSec_CAF1 7624 99 + 1 UCCUCCAAUUAAGAAUGCAAUCAAGGCAGCACCUUCAGUGAAUGCCCCAUUUACAACUGAGUACGCGGCACUUCUUCGGCUGUGCCGCGAGGUGGACAA (((.((.........(((....((((.....))))..(((((((...)))))))......)))((((((((..(....)..)))))))).)).)))... ( -33.90) >DroSim_CAF1 8227 99 + 1 UCCUCCAAUUAAGAAUGCAACCAAGGCAGCACCUUCAGUGAAUGCCCCAUUUACAACUGAGUACGCGGCACUUCUUCGGCUGUGCCGCGAGGUGGACAA (((.((.........(((....((((.....))))..(((((((...)))))))......)))((((((((..(....)..)))))))).)).)))... ( -33.90) >DroEre_CAF1 7383 96 + 1 UACUCCCACUAAGAAUGCAAACGAGGCACCACUAACA---AAUGCCCCUUUUACAACUGAGUACGCGGCACUUUUGAGGCUGUGCCGCGAGGUGGACAA .....(((((.....((.(((.(.((((.........---..)))).).))).))........((((((((..........)))))))).))))).... ( -27.60) >DroYak_CAF1 7761 99 + 1 CACUCCCAUUAUGAAUGCAAUUGAGGCAUCACCAGCAGCGAAUGCCCCUUUUACAACUGAGUACGCGGCACUUUUGAGGCUGUGCCGCGAGGUGGACAA .....(((((......(((.(((..((.......))..))).)))..................((((((((..........)))))))).))))).... ( -27.90) >DroAna_CAF1 8088 90 + 1 ---------GGAAAAGCCAACGGAAAAAUCACCUCCAGCUAGGGCGCCCUUCAACCCCGAGUAUGCGGCAUUCCUGAAGCUCUGUCUCGAGGUGGACAA ---------((.....))..........(((((((.((.((((((...........(((......)))..........)))))).)).))))))).... ( -23.80) >consensus UCCUCCAAUUAAGAAUGCAACCAAGGCAGCACCAUCAGUGAAUGCCCCUUUUACAACUGAGUACGCGGCACUUCUGAGGCUGUGCCGCGAGGUGGACAA .............................((((..................(((......)))((((((((..........)))))))).))))..... (-17.28 = -17.70 + 0.42)

| Location | 6,485,227 – 6,485,326 |
|---|---|
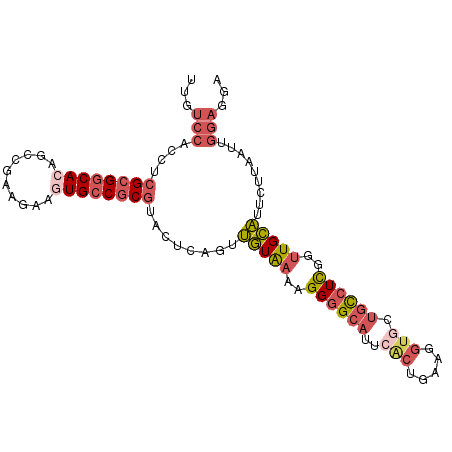
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -20.60 |
| Energy contribution | -22.02 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
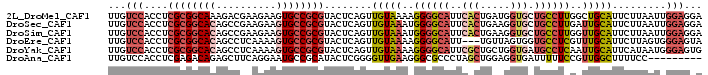
>2L_DroMel_CAF1 6485227 99 - 22407834 UUGUCCACCUCGCGGCAAAGACGAAGAAGUGCCGCGUACUCAGUUGUAAAAGGGGCAUUCACUGAUGGUGCUGCCUUGGCUGCAUUCUUAAUUGGAGGA ((.((((((((((((((............)))))).(((......)))...)))).......(((..((((.((....)).))))..)))..)))).)) ( -29.70) >DroSec_CAF1 7624 99 - 1 UUGUCCACCUCGCGGCACAGCCGAAGAAGUGCCGCGUACUCAGUUGUAAAUGGGGCAUUCACUGAAGGUGCUGCCUUGAUUGCAUUCUUAAUUGGAGGA .......(((((((((((..........))))))).....((((((..(((((((((..(((.....))).)))))......))))..)))))))))). ( -34.10) >DroSim_CAF1 8227 99 - 1 UUGUCCACCUCGCGGCACAGCCGAAGAAGUGCCGCGUACUCAGUUGUAAAUGGGGCAUUCACUGAAGGUGCUGCCUUGGUUGCAUUCUUAAUUGGAGGA .......(((((((((((..........))))))).........(((((..((((((..(((.....))).))))))..)))))..........)))). ( -34.20) >DroEre_CAF1 7383 96 - 1 UUGUCCACCUCGCGGCACAGCCUCAAAAGUGCCGCGUACUCAGUUGUAAAAGGGGCAUU---UGUUAGUGGUGCCUCGUUUGCAUUCUUAGUGGGAGUA ...(((....((((((((..........))))))))((((.((.((((((.((((((((---.......)))))))).))))))..)).)))))))... ( -37.90) >DroYak_CAF1 7761 99 - 1 UUGUCCACCUCGCGGCACAGCCUCAAAAGUGCCGCGUACUCAGUUGUAAAAGGGGCAUUCGCUGCUGGUGAUGCCUCAAUUGCAUUCAUAAUGGGAGUG .....(((((((((((((..........))))))).........(((((..(((((((.(((.....))))))))))..)))))........))).))) ( -35.60) >DroAna_CAF1 8088 90 - 1 UUGUCCACCUCGAGACAGAGCUUCAGGAAUGCCGCAUACUCGGGGUUGAAGGGCGCCCUAGCUGGAGGUGAUUUUUCCGUUGGCUUUUCC--------- ..((((((((((((.....((..((....))..))...))))))))....))))(((..(((.(((((.....)))))))))))......--------- ( -26.30) >consensus UUGUCCACCUCGCGGCACAGCCGAAGAAGUGCCGCGUACUCAGUUGUAAAAGGGGCAUUCACUGAAGGUGCUGCCUCGGUUGCAUUCUUAAUUGGAGGA ...(((....((((((((..........))))))))........(((((..((((((..(((.....))).))))))..))))).........)))... (-20.60 = -22.02 + 1.42)
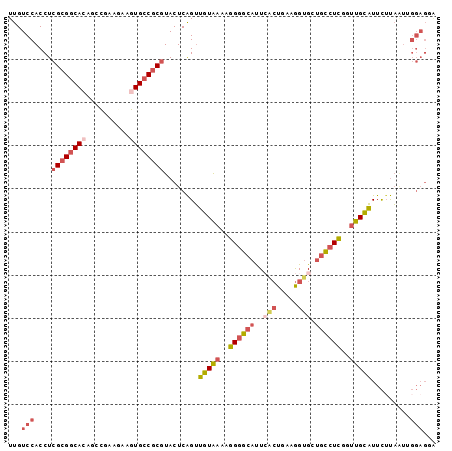
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:50 2006