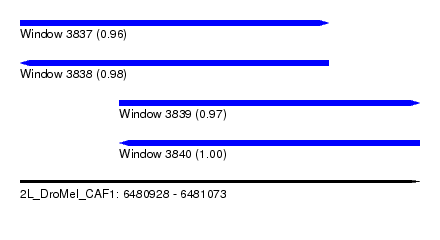
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,480,928 – 6,481,073 |
| Length | 145 |
| Max. P | 0.999828 |

| Location | 6,480,928 – 6,481,040 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -19.68 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6480928 112 + 22407834 UUGACAAUUCUGAGGCAUGAGUACAAACAAACACUCACGCAGUGCACUCUGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUA-UAUACAGUAUUCAGUGCUAGUG .............(((((((((((......((.(((.(((((......))))..).))).)).....((((((((((....)))))))))-).....))))))).)))).... ( -34.30) >DroPse_CAF1 9424 93 + 1 AAAACAAAUGCAAAGCAACAAUACAAACACACGCACAGGCAAUUC-------AAGCAGGCGUAAUAUAUAUGG------AUCGUCAUAUAAUAGAGAGCGUUCAGU------- ......(((((...................((((....((.....-------..))..))))....(((((((------....))))))).......)))))....------- ( -14.70) >DroSec_CAF1 3341 112 + 1 UUGACAAUUCUGAGGCAUGAGCACAAACAAACACUCACGCAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUA-UAUACAGUAUUCAGUGCUAGUG ...............(((.(((((............((((..(((.........))).)))).....((((((((((....)))))))))-)............))))).))) ( -29.10) >DroSim_CAF1 3636 112 + 1 UUGACAAUUCUGAGACAUGAGCACAAACAAACACUCACGCAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUA-UAUACAGUAUUCAGUGCUAGUG ...............(((.(((((............((((..(((.........))).)))).....((((((((((....)))))))))-)............))))).))) ( -29.40) >DroEre_CAF1 3373 112 + 1 UUGACAAUUGUGAGGCAUGAAUACAAACAAACACUCACGCAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAGAAUUCCCACAUA-UAUAUAGUAUUCAGUGCCAGUG .............(((((((((((......((.(((..((..(((.....))).))))).)).(((.(((((((((......))))))))-).))).))))))).)))).... ( -35.70) >DroYak_CAF1 3326 108 + 1 UUGACAAUUGUGAGGAAUGAAUACA----AACACUCACGUAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAGAAUUCCCACAUA-UAUAUAGUAUUCAGUGCCAGUG .............((..(((((((.----.((.(((.(.((.((....)).)).).))).)).(((.(((((((((......))))))))-).))).)))))))...)).... ( -33.80) >consensus UUGACAAUUCUGAGGCAUGAAUACAAACAAACACUCACGCAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUA_UAUACAGUAUUCAGUGCUAGUG .............(((((((((((............((((..(((.........))).))))......((((((((......)))))))).......))))))).)))).... (-19.68 = -21.43 + 1.75)

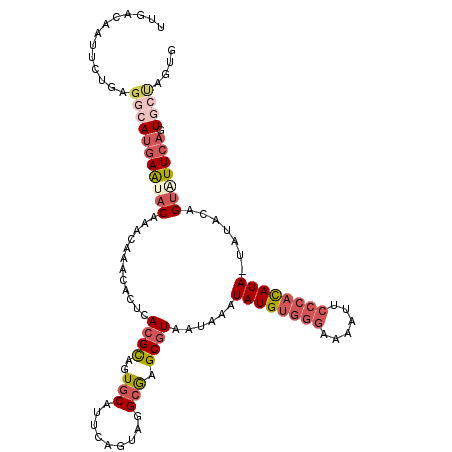

| Location | 6,480,928 – 6,481,040 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -18.47 |
| Energy contribution | -20.11 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6480928 112 - 22407834 CACUAGCACUGAAUACUGUAUA-UAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACAGAGUGCACUGCGUGAGUGUUUGUUUGUACUCAUGCCUCAGAAUUGUCAA ........((((.....(((.(-(((((((((....)))))))))).)))..(((((.......)))))....(((((((((........))))))))).))))......... ( -35.40) >DroPse_CAF1 9424 93 - 1 -------ACUGAACGCUCUCUAUUAUAUGACGAU------CCAUAUAUAUUACGCCUGCUU-------GAAUUGCCUGUGCGUGUGUUUGUAUUGUUGCUUUGCAUUUGUUUU -------...((((((.......((((((.....------.))))))....(((((.((..-------.....))..).))))))))))....(((......)))........ ( -15.00) >DroSec_CAF1 3341 112 - 1 CACUAGCACUGAAUACUGUAUA-UAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUGCGUGAGUGUUUGUUUGUGCUCAUGCCUCAGAAUUGUCAA ((..(((((.(((((..(((.(-(((((((((....)))))))))).))).((((((((((..((....)).)).)))))))).))))))))))..))............... ( -32.50) >DroSim_CAF1 3636 112 - 1 CACUAGCACUGAAUACUGUAUA-UAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUGCGUGAGUGUUUGUUUGUGCUCAUGUCUCAGAAUUGUCAA ((..(((((.(((((..(((.(-(((((((((....)))))))))).))).((((((((((..((....)).)).)))))))).))))))))))..))............... ( -32.70) >DroEre_CAF1 3373 112 - 1 CACUGGCACUGAAUACUAUAUA-UAUGUGGGAAUUCUCCCACAUAUUUAUUACGCUCGCCUACUGAAUGCACUGCGUGAGUGUUUGUUUGUAUUCAUGCCUCACAAUUGUCAA ....((((.(((((((.(((.(-(((((((((....)))))))))).))).((((((((.((.((....)).)).))))))))......)))))))))))............. ( -38.70) >DroYak_CAF1 3326 108 - 1 CACUGGCACUGAAUACUAUAUA-UAUGUGGGAAUUCUCCCACAUAUUUAUUACGCUCGCCUACUGAAUGCACUACGUGAGUGUU----UGUAUUCAUUCCUCACAAUUGUCAA ....((...(((((((.(((.(-(((((((((....)))))))))).))).((((((((.((.((....)).)).)))))))).----.)))))))..))............. ( -33.20) >consensus CACUAGCACUGAAUACUGUAUA_UAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUGCGUGAGUGUUUGUUUGUACUCAUGCCUCACAAUUGUCAA .....(((.(((.(((.......(((((((((....)))))))))......((((((((................))))))))......))).)))))).............. (-18.47 = -20.11 + 1.64)

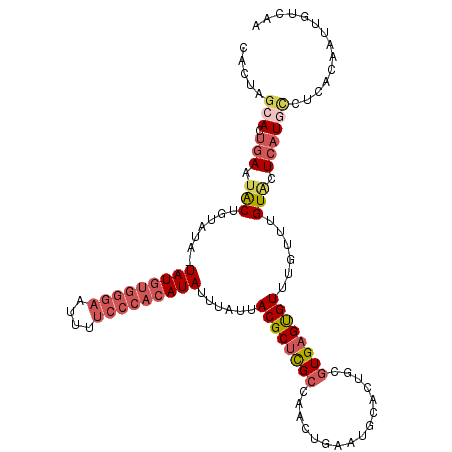
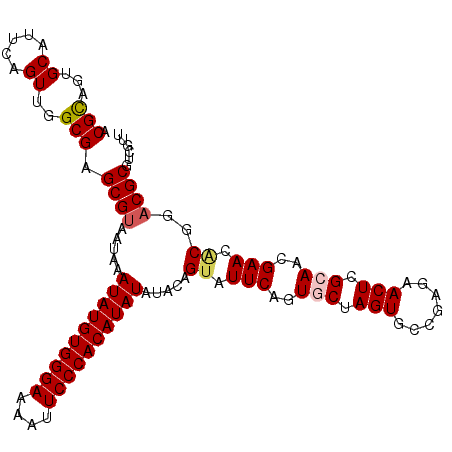
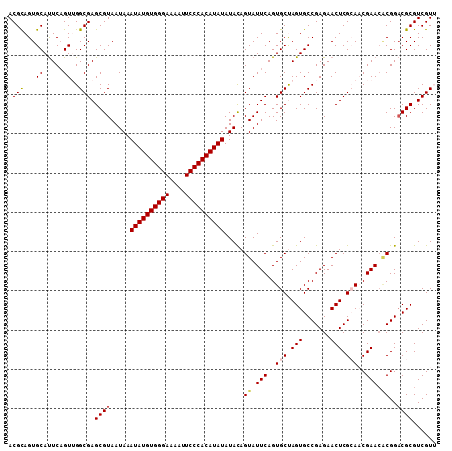
| Location | 6,480,964 – 6,481,073 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.49 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -29.04 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6480964 109 + 22407834 ACGCAGUGCACUCUGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACCCGGUCGCGUCGUU ..((((......))))..((((.((......((((((((((....))))))))))........(((..(((.(((.......))).)))..)))..)).))))...... ( -33.10) >DroSec_CAF1 3377 109 + 1 ACGCAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACGCGGACGCGUCGUU ((((..(((.(((.((((.((((........((((((((((....)))))))))).((((((((...))))).))).......))))))))))).)))...)))).... ( -34.90) >DroSim_CAF1 3672 109 + 1 ACGCAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACGCGGACGCGUCGUU ((((..(((.(((.((((.((((........((((((((((....)))))))))).((((((((...))))).))).......))))))))))).)))...)))).... ( -34.90) >DroEre_CAF1 3409 109 + 1 ACGCAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAGAAUUCCCACAUAUAUAUAGUAUUCAGUGCCAGUGCCGAGAACUCGGACCGAAAACGGACGCGUCGUG .(((..(((........((((((..(.(((.(((((((((......))))))))).))).)..)))...))).((.((((....)))).(((....))))))))..))) ( -35.00) >DroYak_CAF1 3358 109 + 1 ACGUAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAGAAUUCCCACAUAUAUAUAGUAUUCAGUGCCAGUGCCGAGAACUCGGAUCGAACACGGACGCGUCGUG ..................((((((((.(((.(((((((((......))))))))).)))........(((......((((....))))......)))..)))).)))). ( -32.90) >consensus ACGCAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACACGGACGCGUCGUU .(((...((.....))..))).((((.....((((((((((....)))))))))).....((.(((..(((.(((.......))).)))..))).))..))))...... (-29.04 = -29.52 + 0.48)

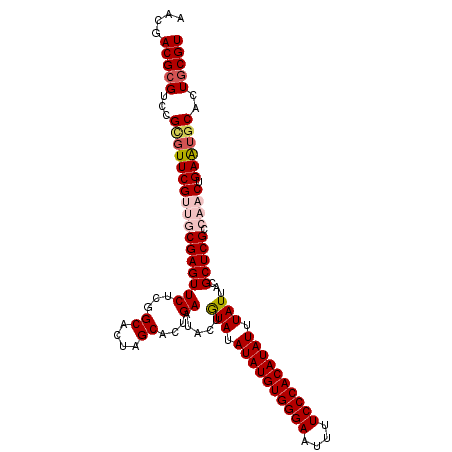
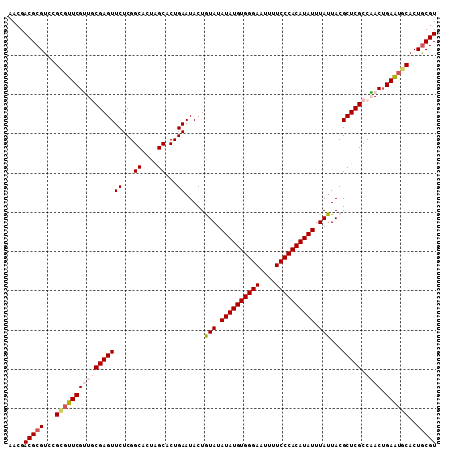
| Location | 6,480,964 – 6,481,073 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.49 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -31.96 |
| Energy contribution | -33.20 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6480964 109 - 22407834 AACGACGCGACCGGGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACAGAGUGCACUGCGU ....(((((...(.(((((((((((((((...((....))...)).....(((.((((((((((....))))))))))....)))))))).)))).)))).)..))))) ( -34.60) >DroSec_CAF1 3377 109 - 1 AACGACGCGUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUGCGU ....(((((...(((((((((((((((((...((....))...)).....(((.((((((((((....))))))))))....)))))))).)))).))))))..))))) ( -41.40) >DroSim_CAF1 3672 109 - 1 AACGACGCGUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUGCGU ....(((((...(((((((((((((((((...((....))...)).....(((.((((((((((....))))))))))....)))))))).)))).))))))..))))) ( -41.40) >DroEre_CAF1 3409 109 - 1 CACGACGCGUCCGUUUUCGGUCCGAGUUCUCGGCACUGGCACUGAAUACUAUAUAUAUGUGGGAAUUCUCCCACAUAUUUAUUACGCUCGCCUACUGAAUGCACUGCGU ....(((((...((.((((((((((....))))....(((...((.....(((.((((((((((....)))))))))).))).....))))).)))))).))..))))) ( -34.10) >DroYak_CAF1 3358 109 - 1 CACGACGCGUCCGUGUUCGAUCCGAGUUCUCGGCACUGGCACUGAAUACUAUAUAUAUGUGGGAAUUCUCCCACAUAUUUAUUACGCUCGCCUACUGAAUGCACUACGU ..(((.(((((.((((..(..((((....))))..)..)))).)).....(((.((((((((((....)))))))))).)))..))))))................... ( -31.60) >consensus AACGACGCGUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUGCGU ....(((((...(((((((((((((((((...((....))...)).....(((.((((((((((....)))))))))).)))...))))).)))).))))))..))))) (-31.96 = -33.20 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:45 2006