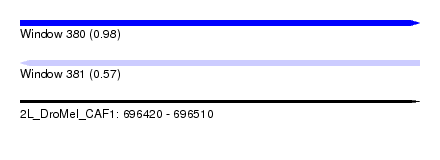
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 696,420 – 696,510 |
| Length | 90 |
| Max. P | 0.978418 |

| Location | 696,420 – 696,510 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 696420 90 + 22407834 AGAGCCCCACUGGGGCCAAACGUCAUAAAUCCACUAAUAAACAGUAACCACAGUAACCCGGCCAGUAU---------GGCCAGUAUGGCGAGUAUGGCA .(.(((((...))))))....((((((...(((..........((.((....)).))..((((.....---------))))....)))....)))))). ( -25.60) >DroSec_CAF1 75457 90 + 1 AGAGUCCCACUGGGGGCAAACGUCAUAAAUCCACUAAUAAACAGUAACCACAGUAACCCGGCCAGUAU---------GGCCAGUGUGGCCAGUAUGGCA ...(((((....)))))....((((((...((((.........((.((....)).))..((((.....---------))))...))))....)))))). ( -28.00) >DroSim_CAF1 80356 99 + 1 AGAGCCCCACUGGGGGCAAACGUCAUAAAUCCACUAAUAAACAGUAACCACAGUAACCCGGCCAGUAUGGCCAGUAUGGCCAGUGUGGCCAGUAUGGCA ...(((((....)))))....((((((.....(((.......)))..............(((((...(((((.....)))))...)))))..)))))). ( -36.70) >DroEre_CAF1 84599 69 + 1 AGAGCCCCACUGGGGGCAAACGUCAUAAAUCCACUAAUAAACAGUAACCACAG---CCC---------------------------GGCCAGUAUGGCA ...(((..(((((((((...................................)---)))---------------------------..)))))..))). ( -17.05) >DroYak_CAF1 80604 69 + 1 AGAGCCCCACUGGGGGCAAACGUCAUAAAUCCACUAAUAAACAGUAACCACGG---CGC---------------------------GGCCAGUAUGGCA ...(((((....)))))....((((((.....(((.......)))......((---(..---------------------------.)))..)))))). ( -19.60) >consensus AGAGCCCCACUGGGGGCAAACGUCAUAAAUCCACUAAUAAACAGUAACCACAGUAACCCGGCCAGUAU_________GGCCAGU_UGGCCAGUAUGGCA ...(((((....)))))....((((((...(((((.......)))..............((((..............)))).....))....)))))). (-17.30 = -17.78 + 0.48)

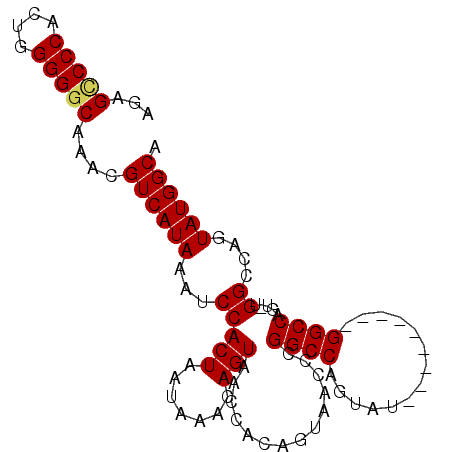
| Location | 696,420 – 696,510 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.58 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 696420 90 - 22407834 UGCCAUACUCGCCAUACUGGCC---------AUACUGGCCGGGUUACUGUGGUUACUGUUUAUUAGUGGAUUUAUGACGUUUGGCCCCAGUGGGGCUCU .(((.((((.((((..((((((---------.....))))))((((.((...((((((.....))))))...))))))...))))...)))).)))... ( -29.90) >DroSec_CAF1 75457 90 - 1 UGCCAUACUGGCCACACUGGCC---------AUACUGGCCGGGUUACUGUGGUUACUGUUUAUUAGUGGAUUUAUGACGUUUGCCCCCAGUGGGACUCU ..((.(((((((((((((((((---------.....)))))).....)))))((((((.....)))))).................))))))))..... ( -28.20) >DroSim_CAF1 80356 99 - 1 UGCCAUACUGGCCACACUGGCCAUACUGGCCAUACUGGCCGGGUUACUGUGGUUACUGUUUAUUAGUGGAUUUAUGACGUUUGCCCCCAGUGGGGCUCU .(((((((((((((...(((((.....)))))...))))))).....))))))(((((.....)))))..............(((((....)))))... ( -38.10) >DroEre_CAF1 84599 69 - 1 UGCCAUACUGGCC---------------------------GGG---CUGUGGUUACUGUUUAUUAGUGGAUUUAUGACGUUUGCCCCCAGUGGGGCUCU .(((.((((((..---------------------------(((---(....((((..(((((....)))))...))))....)))))))))).)))... ( -23.80) >DroYak_CAF1 80604 69 - 1 UGCCAUACUGGCC---------------------------GCG---CCGUGGUUACUGUUUAUUAGUGGAUUUAUGACGUUUGCCCCCAGUGGGGCUCU .(((.((((((..---------------------------(((---.(((.(((((((.....))))).....)).)))..)))..)))))).)))... ( -21.20) >consensus UGCCAUACUGGCCA_ACUGGCC_________AUACUGGCCGGGUUACUGUGGUUACUGUUUAUUAGUGGAUUUAUGACGUUUGCCCCCAGUGGGGCUCU .(((.((((((.......((((..............))))((((...(((.(((((((.....))))).....)).)))...)))))))))).)))... (-16.14 = -17.58 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:53 2006