
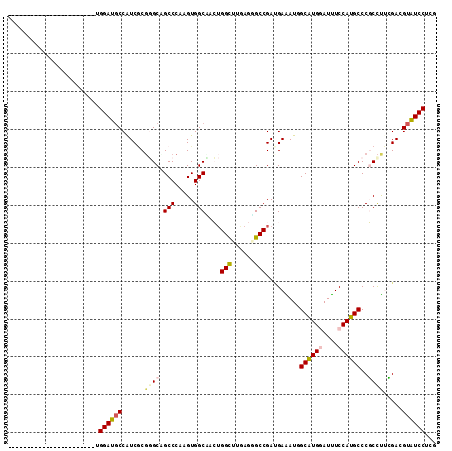
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,446,173 – 6,446,286 |
| Length | 113 |
| Max. P | 0.971570 |

| Location | 6,446,173 – 6,446,286 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6446173 113 + 22407834 UACAUAUAUAAAGCAUUCAUACCUGGAUGCCAUCCCUGGCAGCCCAAGUGGCAACUGGUUUGAGUGCCGAUGAGAUCGCAUGGAUCUCCAUGCCCGCUUGCGACGUGUCCUCG .((((.......(((((((.(((.((.((((((...(((....))).)))))).))))).)))))))((.((((...((((((....))))))...))))))..))))..... ( -38.90) >DroPse_CAF1 24569 90 + 1 -----------------------UGGAUGCCAUCGCGGGCAGCCCAAGUGGCCACUGGCUUGAGGGCCGACGAAAUGGCAUGGAUUUCCAUGCCCGCCUUCGAGGUAUCCUCG -----------------------.(((((((.(((.((((.((((....((((...))))...)))).........(((((((....))))))).)))).))))))))))... ( -47.00) >DroGri_CAF1 27011 90 + 1 -----------------------UGGAUGCCAUCGCGUGCUGCCCAGGUGGCGCAUGGCUUGAGUGCCGAUGAAAUUGCAUGUAUUUCCAUGCCCUCCUUGGACGUAUCCUCG -----------------------.((((((((((..((((..(....)..))))..(((......))))))).....(((((......)))))((.....))..))))))... ( -34.20) >DroMoj_CAF1 39467 90 + 1 -----------------------UGGAUGCCGUCGCGAGCCGCCCAGGUGGCAUAGGGCUUCAGGGCCGAUGAGAUAGCAUGGAUUUCCAUGCCCUCCCUAGACGUGUCCUCG -----------------------.((((((.(((....(((((....))))).(((((((....))))...(((...((((((....)))))).))).))))))))))))... ( -38.40) >DroAna_CAF1 13936 90 + 1 -----------------------UGGAUUCCGUCACGGGCCGCCCACGUGGCUACUGGCUUCAGAGCCGAAGAGAUGGCGUGAAUCUCCAUGCCCGCCUGCGAGGUGUCCUCG -----------------------.((((.((.((.(((((........(((((.(((....)))))))).......((((((......)))))).))))).)))).))))... ( -35.90) >DroPer_CAF1 24933 90 + 1 -----------------------UGGAUGCCAUCGCGGGCAGCCCAUGUGGCCACUGGCUUGAGGGCCGACGAAAUGGCAUGGAUUUCCAUGCCCGCCUUCGAGGUAUCCUCG -----------------------.(((((((.(((.((((.((((....((((...))))...)))).........(((((((....))))))).)))).))))))))))... ( -47.00) >consensus _______________________UGGAUGCCAUCGCGGGCAGCCCAAGUGGCAACUGGCUUGAGGGCCGAUGAAAUGGCAUGGAUUUCCAUGCCCGCCUUCGACGUAUCCUCG ........................((((((......((((.(((.....)))....(((......))).........((((((....))))))..)))).....))))))... (-23.07 = -23.68 + 0.61)


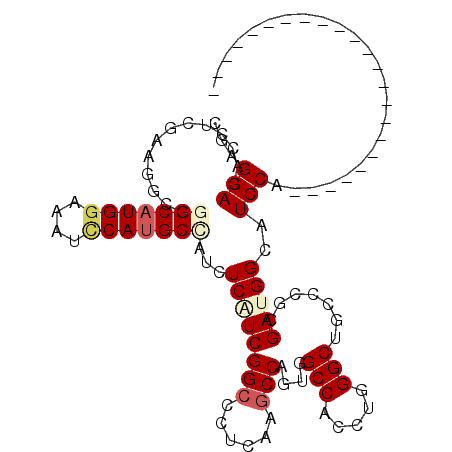
| Location | 6,446,173 – 6,446,286 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -22.79 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6446173 113 - 22407834 CGAGGACACGUCGCAAGCGGGCAUGGAGAUCCAUGCGAUCUCAUCGGCACUCAAACCAGUUGCCACUUGGGCUGCCAGGGAUGGCAUCCAGGUAUGAAUGCUUUAUAUAUGUA .((((.(.((.(....))).((((((....))))))).))))...((((.(((.(((...((((((((((....)))))..)))))....))).))).))))........... ( -40.80) >DroPse_CAF1 24569 90 - 1 CGAGGAUACCUCGAAGGCGGGCAUGGAAAUCCAUGCCAUUUCGUCGGCCCUCAAGCCAGUGGCCACUUGGGCUGCCCGCGAUGGCAUCCA----------------------- ...((((.(((((..(((.(((((((....)))))))........(((((....(((...))).....))))))))..))).)).)))).----------------------- ( -42.00) >DroGri_CAF1 27011 90 - 1 CGAGGAUACGUCCAAGGAGGGCAUGGAAAUACAUGCAAUUUCAUCGGCACUCAAGCCAUGCGCCACCUGGGCAGCACGCGAUGGCAUCCA----------------------- ...((((..(((((.((.(((((((......))))).........(((......))).....)).))))))).....((....)))))).----------------------- ( -31.00) >DroMoj_CAF1 39467 90 - 1 CGAGGACACGUCUAGGGAGGGCAUGGAAAUCCAUGCUAUCUCAUCGGCCCUGAAGCCCUAUGCCACCUGGGCGGCUCGCGACGGCAUCCA----------------------- ...(((..((((..((((.(((((((....))))))).))))..(((((.....((((..........))))))).)).))))...))).----------------------- ( -37.00) >DroAna_CAF1 13936 90 - 1 CGAGGACACCUCGCAGGCGGGCAUGGAGAUUCACGCCAUCUCUUCGGCUCUGAAGCCAGUAGCCACGUGGGCGGCCCGUGACGGAAUCCA----------------------- ...(((..((((((.(((.(.(((((((((.......)))))...(((((((....))).)))).))).).).))).)))).))..))).----------------------- ( -34.60) >DroPer_CAF1 24933 90 - 1 CGAGGAUACCUCGAAGGCGGGCAUGGAAAUCCAUGCCAUUUCGUCGGCCCUCAAGCCAGUGGCCACAUGGGCUGCCCGCGAUGGCAUCCA----------------------- ...((((.(((((..(((.(((((((....)))))))........(((((....(((...))).....))))))))..))).)).)))).----------------------- ( -42.00) >consensus CGAGGACACCUCGAAGGCGGGCAUGGAAAUCCAUGCCAUCUCAUCGGCCCUCAAGCCAGUGGCCACCUGGGCUGCCCGCGAUGGCAUCCA_______________________ ...(((.............(((((((....)))))))...((((((((......)))....(((.....))).......)))))..)))........................ (-22.79 = -23.65 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:31 2006