
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,445,262 – 6,445,369 |
| Length | 107 |
| Max. P | 0.854459 |

| Location | 6,445,262 – 6,445,369 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
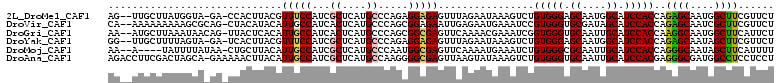
>2L_DroMel_CAF1 6445262 107 + 22407834 AG--UUGCUUAUGGUA-GA-CCACUUACGUUUCCAUCGCUCAUGCCCAGAGGAGAGUUUAGAAUAAAGUCUGUGGGAGCAAUGGCAUCCACCAGAGCAAUGGCUUCGUUCU ..--.......(((..-..-)))...(((...((((.((((.(((((...(.(((.((((...)))).))).).)).))).(((......))))))).))))...)))... ( -27.30) >DroVir_CAF1 28898 108 + 1 CA--AAAAAAAAAGCGCAG-CUACAUACAUUGCCAUCACUCAUGCCCAGCGGAGAAUUGAGAAUGAAAUCCGUGGGUGCGAUAGCAUCCACCAGAGCAAUCGCUUCGUUCU ..--.......(((((..(-((.....((((..((...(((.(((...))))))...))..))))......((((((((....))))))))...)))...)))))...... ( -28.10) >DroGri_CAF1 26130 108 + 1 AA--AUGCUUAAAUAACAG-UUACUCACAUUGCCAUCACUCAUGCCCAGCGGCGAGUUCAAAACGAAAUCGGUGGGUGCAAUUGCAUCCACCAAGGCAAUGGCUUCAUUCU ..--...............-.......(((((((...((((..(((....))))))).............(((((((((....)))))))))..))))))).......... ( -35.30) >DroYak_CAF1 14267 107 + 1 GG--UUGCUUUUAGUA-GA-UCACUUACGUUUCCAUCGCUCAUGCCCAGAGGAGAGUUUAGAAUAAAGUCUGUGGGAGCAAUGGCAUCCACCAGAGCAAUAGCUUCGUUCU ((--.((((....(((-(.-....))))((((((((..(((.......))).(((.((((...)))).)))))))))))...)))).))..(..(((....)))..).... ( -24.30) >DroMoj_CAF1 38598 104 + 1 AA--A----UAUUUUAUAA-CUGCUUACAUUGCCAUCGCUCAUGCCCAAUGGCGAGUUCAAAAUGAAAUCUGUGGGCGCAAUUGCAUCCACCAGGGCAAUAGCUUCAUUUU ..--.----..........-..(((...((((((.((((.(((.....)))))))..............((((((..((....))..))).))))))))))))........ ( -27.30) >DroAna_CAF1 13136 110 + 1 AGACCUUCGACUAGCA-GAAAAACUUACAUUGCCAUCGCUCAUGCCAAGGGGCGAGUUAAGUAUAAAGUCUGUGGGUGCAAUUGCAUCCACGAGGGCGAUGGCCUCCUCCU .....(((........-)))...........((((((((((.((((....))))................(((((((((....))))))))).))))))))))........ ( -38.60) >consensus AA__AUGCUUAUAGUA_AA_CUACUUACAUUGCCAUCGCUCAUGCCCAGAGGAGAGUUCAGAAUAAAAUCUGUGGGUGCAAUUGCAUCCACCAGAGCAAUGGCUUCGUUCU .............................(((((...((....)).....)))))................(((((.((....)).)))))..((((....))))...... (-15.38 = -15.02 + -0.36)

| Location | 6,445,262 – 6,445,369 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -18.53 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6445262 107 - 22407834 AGAACGAAGCCAUUGCUCUGGUGGAUGCCAUUGCUCCCACAGACUUUAUUCUAAACUCUCCUCUGGGCAUGAGCGAUGGAAACGUAAGUGG-UC-UACCAUAAGCAA--CU ............(((((.((((((((.(((((((((((.((((..................))))))...)))))))))..((....)).)-))-)))))..)))))--.. ( -34.27) >DroVir_CAF1 28898 108 - 1 AGAACGAAGCGAUUGCUCUGGUGGAUGCUAUCGCACCCACGGAUUUCAUUCUCAAUUCUCCGCUGGGCAUGAGUGAUGGCAAUGUAUGUAG-CUGCGCUUUUUUUUU--UG ((((.((((((...(((...(((.((((((((((.((((((((...............)))).)))).....)))))))).)).)))..))-)..)))))).)))).--.. ( -32.76) >DroGri_CAF1 26130 108 - 1 AGAAUGAAGCCAUUGCCUUGGUGGAUGCAAUUGCACCCACCGAUUUCGUUUUGAACUCGCCGCUGGGCAUGAGUGAUGGCAAUGUGAGUAA-CUGUUAUUUAAGCAU--UU .(((((....((((((((((((((.(((....))).)))))))...........(((((((....)))..))))...)))))))((((((.-....))))))..)))--)) ( -34.60) >DroYak_CAF1 14267 107 - 1 AGAACGAAGCUAUUGCUCUGGUGGAUGCCAUUGCUCCCACAGACUUUAUUCUAAACUCUCCUCUGGGCAUGAGCGAUGGAAACGUAAGUGA-UC-UACUAAAAGCAA--CC ............(((((.((((((((.(((((((((((.((((..................))))))...)))))))))..((....)).)-))-)))))..)))))--.. ( -31.87) >DroMoj_CAF1 38598 104 - 1 AAAAUGAAGCUAUUGCCCUGGUGGAUGCAAUUGCGCCCACAGAUUUCAUUUUGAACUCGCCAUUGGGCAUGAGCGAUGGCAAUGUAAGCAG-UUAUAAAAUA----U--UU ........((((((((....((((.(((....))).)))).((.(((.....))).))(((....)))....))))))))...........-..........----.--.. ( -27.40) >DroAna_CAF1 13136 110 - 1 AGGAGGAGGCCAUCGCCCUCGUGGAUGCAAUUGCACCCACAGACUUUAUACUUAACUCGCCCCUUGGCAUGAGCGAUGGCAAUGUAAGUUUUUC-UGCUAGUCGAAGGUCU ((.(((((((((((((....((((.(((....))).))))..................(((....)))....))))))))(((....)))))))-).))((.(....).)) ( -33.40) >consensus AGAACGAAGCCAUUGCCCUGGUGGAUGCAAUUGCACCCACAGACUUCAUUCUAAACUCGCCGCUGGGCAUGAGCGAUGGCAAUGUAAGUAG_UC_UACUAUAAGCAU__CU ........((((((((....((((.(((....))).))))...............(((......))).....))))))))............................... (-18.53 = -18.78 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:29 2006