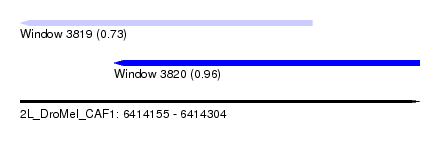
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,414,155 – 6,414,304 |
| Length | 149 |
| Max. P | 0.963576 |

| Location | 6,414,155 – 6,414,264 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.01 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6414155 109 - 22407834 AGGAAAACU------GGUCAGCCAAAAUUAAUAAGACUCAGAUGGCCAGUAUUCCUGGCCAAUUAAAACGGAAACGGAUUUCAAUGUGUCUUUCAAAGACGUGCUCUUUACAGGA .......((------(...(((..........(((((.((..(((((((.....))))))).......((....))........)).)))))..........))).....))).. ( -27.65) >DroEre_CAF1 1265 97 - 1 AGCGA-------AAAAGUCAGGCAGUAAAAA-----------UGGCCAGUGUUCCUGGCCAGUUAAAACGGAAACGGGUUUCAAUGUAUCUGGCAACGAACUGCUCUCUACAGGA .....-------...((...((((((.....-----------(((((((.....)))))))(((....((((.(((........))).))))..)))..))))))..))...... ( -25.90) >DroYak_CAF1 972 115 - 1 AGCGAAACUCGAAAAAGGCAGACAGUAAAAAUAUGAUUUAGAUGGCCCGUGUUGCUGGCCAUUUGAAACGGCAACGGAUUUUAAUAAAUUCCUCAACGACCUGCUCUUCACAGGA ........(((....(((.......(((((......(((((((((((.(.....).))))))))))).((....))..))))).......)))...)))((((.......)))). ( -27.34) >consensus AGCGAAACU___AAAAGUCAGACAGUAAAAAUA_GA_U_AGAUGGCCAGUGUUCCUGGCCAAUUAAAACGGAAACGGAUUUCAAUGUAUCUCUCAACGACCUGCUCUUUACAGGA ...............((.(((.....................(((((((.....))))))).......((....))........................))))).......... (-15.82 = -16.27 + 0.45)


| Location | 6,414,190 – 6,414,304 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.95 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -16.26 |
| Energy contribution | -17.48 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6414190 114 - 22407834 AUAGACUGGAACAUUAGUUUACAGAUAAAGUCUUUAAGAUAGGAAAACU------GGUCAGCCAAAAUUAAUAAGACUCAGAUGGCCAGUAUUCCUGGCCAAUUAAAACGGAAACGGAUU .....(((((((....)))).)))....(((((((((..((((((.(((------(((((......................)))))))).)))))).....))))).((....)))))) ( -27.65) >DroEre_CAF1 1300 102 - 1 AUAGACUGGAACUUUGGUUUACAGAUAAAGACUUUAAGAUAGCGA-------AAAAGUCAGGCAGUAAAAA-----------UGGCCAGUGUUCCUGGCCAGUUAAAACGGAAACGGGUU ....((((((((....)))).........((((((..........-------.))))))...)))).....-----------(((((((.....))))))).......((....)).... ( -25.70) >DroYak_CAF1 1007 119 - 1 AUAAAAUGGAACAUUGGU-UACAGAUAAAGUCUUGAAGAUAGCGAAACUCGAAAAAGGCAGACAGUAAAAAUAUGAUUUAGAUGGCCCGUGUUGCUGGCCAUUUGAAACGGCAACGGAUU ......((.(((....))-).))......((((.(.......((.....)).......))))).............(((((((((((.(.....).))))))))))).((....)).... ( -24.24) >consensus AUAGACUGGAACAUUGGUUUACAGAUAAAGUCUUUAAGAUAGCGAAACU___AAAAGUCAGACAGUAAAAAUA_GA_U_AGAUGGCCAGUGUUCCUGGCCAAUUAAAACGGAAACGGAUU .....(((((((....)))).))).....(((((....................))))).......................(((((((.....))))))).......((....)).... (-16.26 = -17.48 + 1.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:26 2006