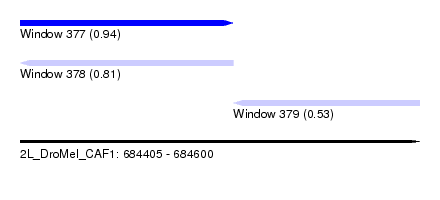
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 684,405 – 684,600 |
| Length | 195 |
| Max. P | 0.944674 |

| Location | 684,405 – 684,509 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -34.10 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 684405 104 + 22407834 UUUCUCUUUUUCGGCCCCACCUGGGCAGGUAUUUCCUUUGGCUGGUAGGCAACGGUAAGCAACACAUGCGAACCGCACCAAAGAACAGCUGAAGGAGGCGAAAG ........(((((((((.....)))).....(((((((..((((...((...((((..(((.....)))..))))..))......))))..)))))))))))). ( -37.40) >DroSim_CAF1 77355 104 + 1 UUUCUCUUUUUCGGCCCCACCUGGGCAGGUAUUUCCUUUGGCUGGUAGGCAACGGUAAGCAACACAUGCGAACCGCACCGAAGAACAGCUAAAGGAGGGAUGAG ..........(((..(((((((....))))...(((((((((((...((...((((..(((.....)))..))))..))......)))))))))))))).))). ( -37.70) >DroEre_CAF1 81565 104 + 1 UUUCUCUUUUUCUGCCCCACCUGGGCAGGUAUUUUCUUUGGCUGGUAGGCAACGGUAAGCAACACAUGCGAACCGCACCAAAGAACAGCUGAAGGAGGGGCAAG ............((((((((((....))))...(((((..((((...((...((((..(((.....)))..))))..))......))))..))))))))))).. ( -39.50) >DroYak_CAF1 77589 104 + 1 UUUCUCUUUUUCGGCCCCACCUGGGCAGGUAUUUCCUUUGACUGGUAGGCAACGGUAAGCAACACAUGCGAACCGCACCAAAGAACAGCUGAAGGAGGGGCAAG .............(((((((((....))))...((((((..(((...((...((((..(((.....)))..))))..))......)))..)))))))))))... ( -35.70) >consensus UUUCUCUUUUUCGGCCCCACCUGGGCAGGUAUUUCCUUUGGCUGGUAGGCAACGGUAAGCAACACAUGCGAACCGCACCAAAGAACAGCUGAAGGAGGGGCAAG .............(((((((((....))))...(((((((((((...((...((((..(((.....)))..))))..))......))))))))))))))))... (-34.10 = -34.60 + 0.50)

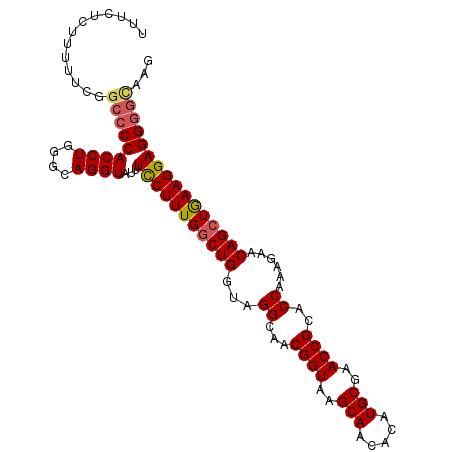
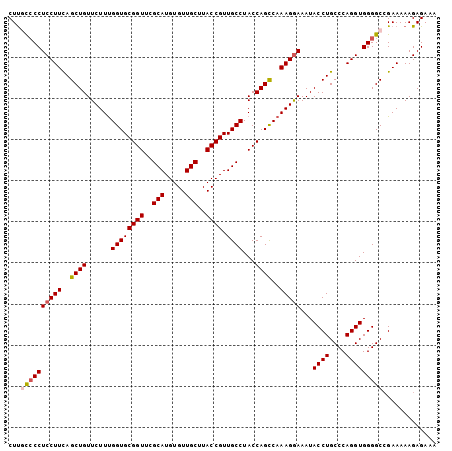
| Location | 684,405 – 684,509 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.93 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.810023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 684405 104 - 22407834 CUUUCGCCUCCUUCAGCUGUUCUUUGGUGCGGUUCGCAUGUGUUGCUUACCGUUGCCUACCAGCCAAAGGAAAUACCUGCCCAGGUGGGGCCGAAAAAGAGAAA .((((((((((((..((((......((((((((..(((.....)))..)))).))))...))))..)))))..(((((....))))).)).)))))........ ( -35.20) >DroSim_CAF1 77355 104 - 1 CUCAUCCCUCCUUUAGCUGUUCUUCGGUGCGGUUCGCAUGUGUUGCUUACCGUUGCCUACCAGCCAAAGGAAAUACCUGCCCAGGUGGGGCCGAAAAAGAGAAA (((.....((((((.((((......((((((((..(((.....)))..)))).))))...)))).))))))....((..(....)..)).........)))... ( -34.20) >DroEre_CAF1 81565 104 - 1 CUUGCCCCUCCUUCAGCUGUUCUUUGGUGCGGUUCGCAUGUGUUGCUUACCGUUGCCUACCAGCCAAAGAAAAUACCUGCCCAGGUGGGGCAGAAAAAGAGAAA .(((((((.(((.(((...((((((((((((((..(((.....)))..))))).........))))))))).....)))...))).)))))))........... ( -39.50) >DroYak_CAF1 77589 104 - 1 CUUGCCCCUCCUUCAGCUGUUCUUUGGUGCGGUUCGCAUGUGUUGCUUACCGUUGCCUACCAGUCAAAGGAAAUACCUGCCCAGGUGGGGCCGAAAAAGAGAAA ...((((((((((..((((......((((((((..(((.....)))..)))).))))...))))..)))))...((((....)))))))))............. ( -36.90) >consensus CUUGCCCCUCCUUCAGCUGUUCUUUGGUGCGGUUCGCAUGUGUUGCUUACCGUUGCCUACCAGCCAAAGGAAAUACCUGCCCAGGUGGGGCCGAAAAAGAGAAA ...((((((((((..((((......((((((((..(((.....)))..)))).))))...))))..)))))...((((....)))))))))............. (-31.30 = -31.93 + 0.63)

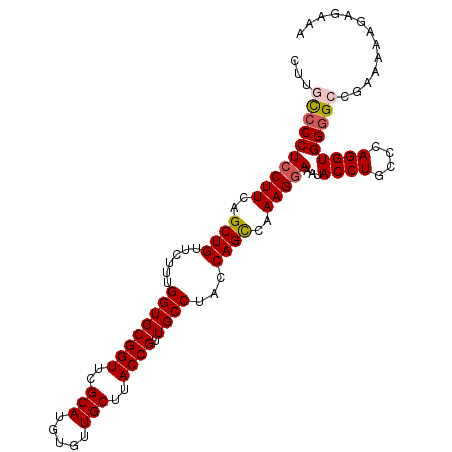
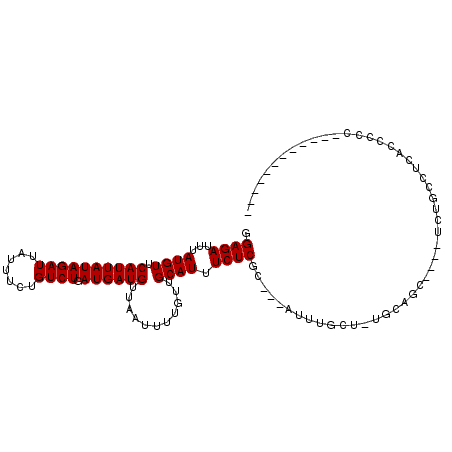
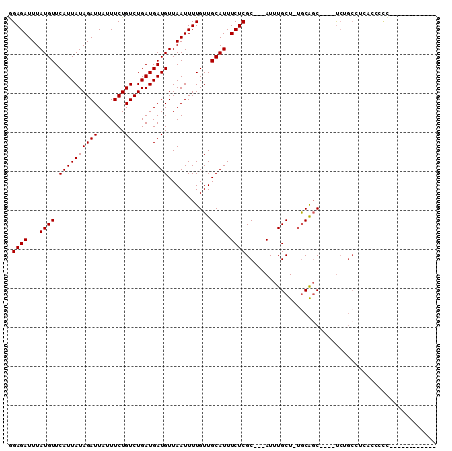
| Location | 684,509 – 684,600 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -19.11 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 684509 91 - 22407834 GGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGC---AUUUGCU-UGCAGC----UCUGCCCCUCCGCC-----------A ((((..(((...((((((((((.....)))))..)))))...)))....((((((......((---....)).-))))))----.......))))...-----------. ( -19.10) >DroSec_CAF1 72569 92 - 1 GGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGC---AUUUGCUUUGCAGC----UCUGNNNNNNNNNN-----------N .((((((.....((((((((((.....)))))..)))))....))))))((((((......((---....))..))))))----..............-----------. ( -15.60) >DroWil_CAF1 45054 98 - 1 AGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGCAGCAUUUGCU-UGCAGCAGAUUCCGCUUCAUCUCUC----------- .((((............(((((.....)))))..(((((.((....(((((((((........(((....)))-)))))))))....)).)))))))))----------- ( -23.20) >DroYak_CAF1 77693 102 - 1 GGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGC---AUUUGCU-UGCAGC----UCUGCUGCGCCCCCCUCUCUCCACCA (((((.(((...((((((((((.....)))))..)))))...))).......((.......((---....)).-.(((((----...))))))).......))))).... ( -23.20) >DroMoj_CAF1 83299 90 - 1 AGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGC---AUUUGCU--GCAGC----CAAGCAACGUCAUU-----------U ...((((((((.....))))))))..........(((((((((..(((.((((((......((---....)))--)))))----.))).)))))))))-----------. ( -18.30) >DroAna_CAF1 38372 94 - 1 GGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGC---AUUUGCU-UACGUC----UCAACCUCAGUCCC--------AGUC (((((((((((.....))))))))....(((..(((.(((((.........(((.......))---)......-.)))))----)))....)))))).--------.... ( -15.29) >consensus GGAGAUUUAUGUUCAUUAUAGAUUAUUUCUGUCUGAUGAUGUUAAUUUUGUUGCAUUUCUCGC___AUUUGCU_UGCAGC____UCUGCCUCACCCCC____________ .((((...((((.((((((((((.......)))).))))))...........)))).))))................................................. (-11.57 = -11.57 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:52 2006