
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
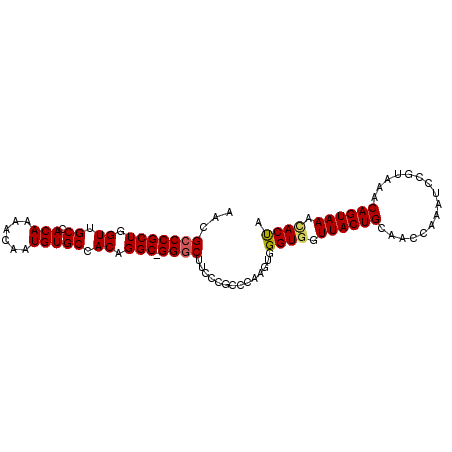
| Location | 6,412,988 – 6,413,083 |
| Length | 95 |
| Max. P | 0.888635 |

| Location | 6,412,988 – 6,413,083 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -23.75 |
| Energy contribution | -23.94 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
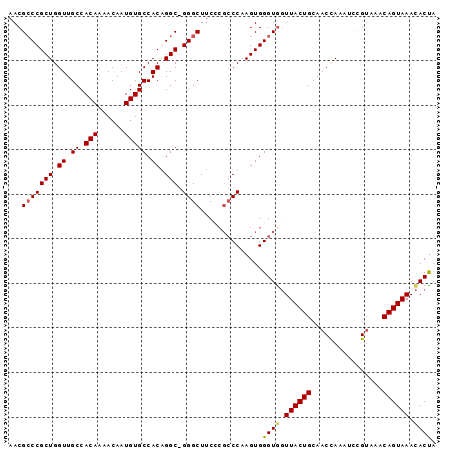
>2L_DroMel_CAF1 6412988 95 + 22407834 AACGGCCGCUGGUUGCCACAAAACAAUGUGCCACAGGC-GGGCUUCCCGCCCAAGUGGGUGCUUACUGCAACCAAAUACGUAAACAGUAAAUACUA .(((.....(((((((((((......))))((((.(((-(((...))))))...)))).........)))))))....)))............... ( -31.20) >DroSec_CAF1 41227 92 + 1 AACGCCCGCUGGUUGCCACAAAACAAUGUGCCACGGGCAGGGCUUC----CCAAGUGGGUGGUUACUGCAACCAAAUCCGUAAACAGUAAACACUA ..((((((((((..((((((......)))(((...)))..)))...----)).)))))))).((((((..((.......))...))))))...... ( -27.80) >DroSim_CAF1 41725 95 + 1 AACGCCCGCUGGUUGCCACAAAACAAUGUGCCACGGGC-GGGCUUCCCGCCCAAGUGGGUAGUUACUGCAACCAAAUCCGAAAACAGUAAACACUA ...(((((((.((.((.(((......))))).))((((-(((...))))))).)))))))..((((((................))))))...... ( -33.69) >DroYak_CAF1 49567 95 + 1 AACGCCCGCUGGUUGCCACAAAACAAUGUGCCACAGGC-GGGCUUCUCGCCCAAGUGGGUGGUUACUGCAACUAGAUCUGUAAACAGUAAACACCA ...(((((((.((.((.(((......))))).)).)))-))))...((((((....))))))((((((..((.......))...))))))...... ( -33.10) >consensus AACGCCCGCUGGUUGCCACAAAACAAUGUGCCACAGGC_GGGCUUCCCGCCCAAGUGGGUGGUUACUGCAACCAAAUCCGUAAACAGUAAACACUA ...(((((((.((.((.(((......))))).)).))).))))..............((((.((((((................)))))).)))). (-23.75 = -23.94 + 0.19)

| Location | 6,412,988 – 6,413,083 |
|---|---|
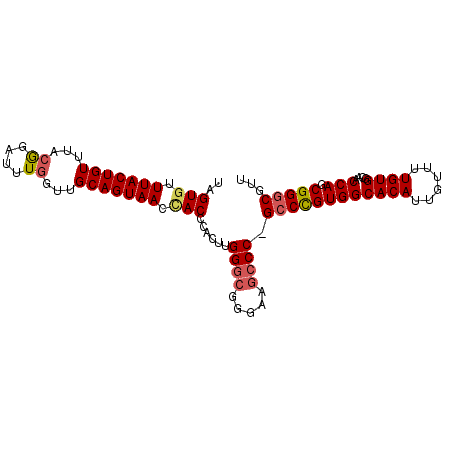
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -26.39 |
| Energy contribution | -27.08 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
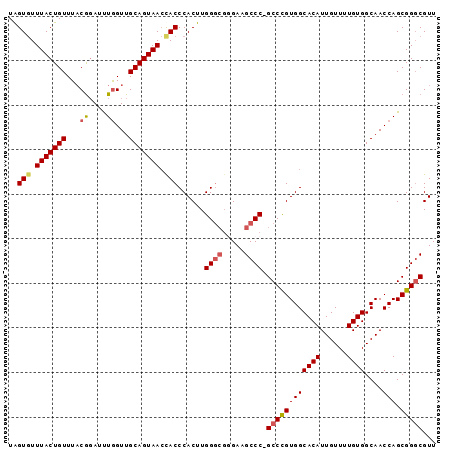
>2L_DroMel_CAF1 6412988 95 - 22407834 UAGUAUUUACUGUUUACGUAUUUGGUUGCAGUAAGCACCCACUUGGGCGGGAAGCCC-GCCUGUGGCACAUUGUUUUGUGGCAACCAGCGGCCGUU ((((....))))....(((...(((((((.........((((..(((((((...)))-))))))))((((......))))))))))))))...... ( -34.60) >DroSec_CAF1 41227 92 - 1 UAGUGUUUACUGUUUACGGAUUUGGUUGCAGUAACCACCCACUUGG----GAAGCCCUGCCCGUGGCACAUUGUUUUGUGGCAACCAGCGGGCGUU ...((.(((((((..((.......)).))))))).))(((....))----)..((((.((...((.((((......)))).))....))))))... ( -29.80) >DroSim_CAF1 41725 95 - 1 UAGUGUUUACUGUUUUCGGAUUUGGUUGCAGUAACUACCCACUUGGGCGGGAAGCCC-GCCCGUGGCACAUUGUUUUGUGGCAACCAGCGGGCGUU ....((((.(((....)))..((((((((.........((((..(((((((...)))-))))))))((((......)))))))))))).))))... ( -36.40) >DroYak_CAF1 49567 95 - 1 UGGUGUUUACUGUUUACAGAUCUAGUUGCAGUAACCACCCACUUGGGCGAGAAGCCC-GCCUGUGGCACAUUGUUUUGUGGCAACCAGCGGGCGUU .((((.(((((((..((.......)).))))))).)))).....((((.....))))-((((((((((((......))))....))).)))))... ( -36.30) >consensus UAGUGUUUACUGUUUACGGAUUUGGUUGCAGUAACCACCCACUUGGGCGGGAAGCCC_GCCCGUGGCACAUUGUUUUGUGGCAACCAGCGGGCGUU ..(((.(((((((...((....))...))))))).)))......((((.....)))).((((((((((((......))))....))).)))))... (-26.39 = -27.08 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:24 2006