
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,410,672 – 6,410,772 |
| Length | 100 |
| Max. P | 0.871181 |

| Location | 6,410,672 – 6,410,772 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.20 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
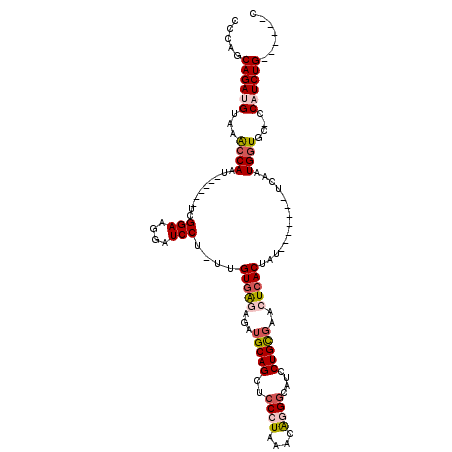
>2L_DroMel_CAF1 6410672 100 + 22407834 CCCAGCAGAUGUAAACCAAU------UCGGAAGGAUCCU-UUGUAAAAGAUGCAGCUCCCUAAACAGGCCAUCCUGCGAACUCACUAU-------UCAAUGAUGCCCCAUCUG------C ....(((((((.......((------((....))))...-..........(((((...(((....))).....)))))..........-------............))))))------) ( -19.50) >DroSec_CAF1 38960 99 + 1 CCCAGCAGAUGUAAACCAAU------UCGGAAGGAUCCG-UUGUGAGAGAUGCAGCUCCCUAAACAGGGCAUCCUGCGAACUCACUAU-------UCAAUGGUGC-CCAUCUG------C ....(((((((...((((..------.((((....))))-..(((((...(((((..((((....))))....)))))..)))))...-------....))))..-.))))))------) ( -35.30) >DroSim_CAF1 39449 100 + 1 CCCAGCAGAUGUAAACCAAU------UCGGAAGGAUCCUUUUGUGAGAGAUGCAGCUCCCUAAACAGGGCAUCCUGCGAACUCACUAU-------UCAAUGGUGC-CCAUCUG------C ....(((((((...((((((------((....))))......(((((...(((((..((((....))))....)))))..)))))...-------....))))..-.))))))------) ( -35.20) >DroEre_CAF1 37554 111 + 1 C-AAGCAGAGGUAAACCAAU------UCGGAAGGAUCCU-UUGUGGAAGGUGCAGCUCCAUAAACGGGGCAUCCUGUGAACUCACUAUUGUGCCGACAAUGGUGC-CCAUCUGCAACUGC .-..(((((((...((((.(------((....)))(((.-....))).((..(((((((......))))).....(((....)))...))..)).....))))..-)).)))))...... ( -38.70) >DroYak_CAF1 42978 111 + 1 C-AAGCAGAGGUAAGCCAAGAGGUAAACGGAAGGAUCCU-UUGUGGGAGGAGCAGCUCCAUAAACAGGGCAUCCUGUGAACUCACUAUAAUGCCCUCAAUGGUGC-CCAUCUG------C .-..(((((((...(((((((((....(....)...)))-))((.((((......))))....))(((((((...(((....)))....)))))))...))))..-)).))))------) ( -39.10) >consensus CCCAGCAGAUGUAAACCAAU______UCGGAAGGAUCCU_UUGUGAGAGAUGCAGCUCCCUAAACAGGGCAUCCUGCGAACUCACUAU_______UCAAUGGUGC_CCAUCUG______C .....((((((...((((..........(((....)))....(((((...(((((..((((....))))....)))))..)))))..............))))....))))))....... (-20.00 = -21.20 + 1.20)

| Location | 6,410,672 – 6,410,772 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -35.69 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6410672 100 - 22407834 G------CAGAUGGGGCAUCAUUGA-------AUAGUGAGUUCGCAGGAUGGCCUGUUUAGGGAGCUGCAUCUUUUACAA-AGGAUCCUUCCGA------AUUGGUUUACAUCUGCUGGG (------((((((((.((((..(((-------((.....)))))...)))).))......(((((....((((((....)-))))).)))))..------.........))))))).... ( -30.80) >DroSec_CAF1 38960 99 - 1 G------CAGAUGG-GCACCAUUGA-------AUAGUGAGUUCGCAGGAUGCCCUGUUUAGGGAGCUGCAUCUCUCACAA-CGGAUCCUUCCGA------AUUGGUUUACAUCUGCUGGG (------((((((.-..((((....-------...(((((...((((....((((....))))..))))....)))))..-((((....)))).------..))))...))))))).... ( -39.00) >DroSim_CAF1 39449 100 - 1 G------CAGAUGG-GCACCAUUGA-------AUAGUGAGUUCGCAGGAUGCCCUGUUUAGGGAGCUGCAUCUCUCACAAAAGGAUCCUUCCGA------AUUGGUUUACAUCUGCUGGG (------((((((.-..((((....-------...(((((...((((....((((....))))..))))....)))))....(((....)))..------..))))...))))))).... ( -36.90) >DroEre_CAF1 37554 111 - 1 GCAGUUGCAGAUGG-GCACCAUUGUCGGCACAAUAGUGAGUUCACAGGAUGCCCCGUUUAUGGAGCUGCACCUUCCACAA-AGGAUCCUUCCGA------AUUGGUUUACCUCUGCUU-G ......(((((.((-..((((...((((.......(((....)))(((((((.(((....))).))....((((.....)-)))))))).))))------..))))...)))))))..-. ( -34.30) >DroYak_CAF1 42978 111 - 1 G------CAGAUGG-GCACCAUUGAGGGCAUUAUAGUGAGUUCACAGGAUGCCCUGUUUAUGGAGCUGCUCCUCCCACAA-AGGAUCCUUCCGUUUACCUCUUGGCUUACCUCUGCUU-G (------((((.((-((.((((..((((((((...(((....)))..))))))))....)))).)).(((..........-.(((....)))...........)))...)))))))..-. ( -37.45) >consensus G______CAGAUGG_GCACCAUUGA_______AUAGUGAGUUCGCAGGAUGCCCUGUUUAGGGAGCUGCAUCUCUCACAA_AGGAUCCUUCCGA______AUUGGUUUACAUCUGCUGGG .......((((((....((((..............(((((((((((((....))))).....))))).)))...........(((....)))..........))))...))))))..... (-21.62 = -21.98 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:20 2006