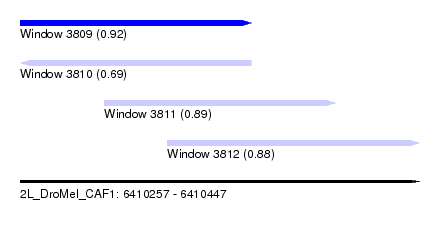
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,410,257 – 6,410,447 |
| Length | 190 |
| Max. P | 0.923752 |

| Location | 6,410,257 – 6,410,367 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -23.38 |
| Energy contribution | -26.26 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6410257 110 + 22407834 AUUCCACGGUAUAUUAAUCUUUUCGAUCGGACAAUCAUUGCAGAUUUCUGCACGUUUGUUUACAUAU------AUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGU .......(((.((((((((.....))).((((((....(((((....)))))...))))))......------.))))))))(((...(((((....)))))....)))....... ( -29.70) >DroSec_CAF1 38546 110 + 1 AUUCCAUGGUAUAUUAAUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCGCGUUUGUUUACAUAU------AUAGUAGCCGGCUUUUGCACUUCGGUGUUCAUUACCACCACGU ....((((((...(..(((.....)))..)...))))))((((....))))((((..(.((((....------...)))).)((.....((((....))))......))...)))) ( -22.70) >DroSim_CAF1 39027 110 + 1 AUUCCACGGUAUAUUAAUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCGCGUUUGUUUACAUAU------AUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGU .......(((.((((((((.....))).((((((..(((((((....)))).)))))))))......------.))))))))(((...(((((....)))))....)))....... ( -29.50) >DroEre_CAF1 37155 115 + 1 AUUCCAUGGCAUAUUAAUCUCUUCGAUCGGACAAUCAUGGCAGAU-UCUGCACGUUUGUUUACAUAUACGUGUAUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGU .......(((.(((((((((((..(((......)))..)).))))-..(((((((.((....))...)))))))))))))))(((....((((....)))).....)))....... ( -32.00) >DroYak_CAF1 42570 110 + 1 AUUCCACGGUAUAUUAAUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCACGUUUGUUUACAUAU------AUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGU .......(((.((((((((.....))).((((((..(((((((....)))).)))))))))......------.))))))))(((....((((....)))).....)))....... ( -27.90) >consensus AUUCCACGGUAUAUUAAUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCACGUUUGUUUACAUAU______AUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGU .......(((.((((((((.....))).((((((.....((((....))))....)))))).............))))))))(((....((((....)))).....)))....... (-23.38 = -26.26 + 2.88)

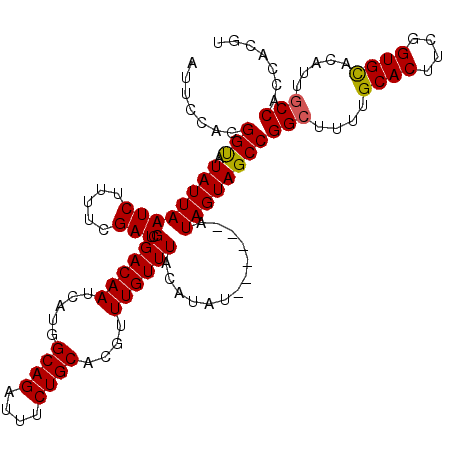
| Location | 6,410,257 – 6,410,367 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6410257 110 - 22407834 ACGUGGUGGCAAUGUGCACCGAAGUGCAAAAGCCGGCUACUAU------AUAUGUAAACAAACGUGCAGAAAUCUGCAAUGAUUGUCCGAUCGAAAAGAUUAAUAUACCGUGGAAU ..((((((((....(((((....)))))...))).)))))...------.(((((..((((.(((((((....)))).))).))))..((((.....)))).)))))......... ( -28.60) >DroSec_CAF1 38546 110 - 1 ACGUGGUGGUAAUGAACACCGAAGUGCAAAAGCCGGCUACUAU------AUAUGUAAACAAACGCGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAUUAAUAUACCAUGGAAU .(((((((.((((......(((.(..(((.....(..(((...------....)))..)....(.((((....)))))....)))..)..))).....))))...))))))).... ( -21.90) >DroSim_CAF1 39027 110 - 1 ACGUGGUGGCAAUGUGCACCGAAGUGCAAAAGCCGGCUACUAU------AUAUGUAAACAAACGCGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAUUAAUAUACCGUGGAAU ..((((((((....(((((....)))))...))).)))))...------.............(((((((....))))..(((((....)))))................))).... ( -27.20) >DroEre_CAF1 37155 115 - 1 ACGUGGUGGCAAUGUGCACCGAAGUGCGAAAGCCGGCUACUAUACACGUAUAUGUAAACAAACGUGCAGA-AUCUGCCAUGAUUGUCCGAUCGAAGAGAUUAAUAUGCCAUGGAAU .((((((((((.(.(((((....(.((....)))(..(((((((....)))).)))..)....))))).)-...)))).(((((....))))).............)))))).... ( -31.60) >DroYak_CAF1 42570 110 - 1 ACGUGGUGGCAAUGUGCACCGAAGUGCGAAAGCCGGCUACUAU------AUAUGUAAACAAACGUGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAUUAAUAUACCGUGGAAU ..((((((((....(((((....)))))...))).)))))...------.(((((..((((.(((((((....))).)))).))))..((((.....)))).)))))......... ( -27.90) >consensus ACGUGGUGGCAAUGUGCACCGAAGUGCAAAAGCCGGCUACUAU______AUAUGUAAACAAACGUGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAUUAAUAUACCGUGGAAU .(((((((((....(((((....)))))...)))((.(((((((....)))).))).((((.(((((((....)))).))).))))))((((.....)))).....)))))).... (-25.14 = -25.42 + 0.28)

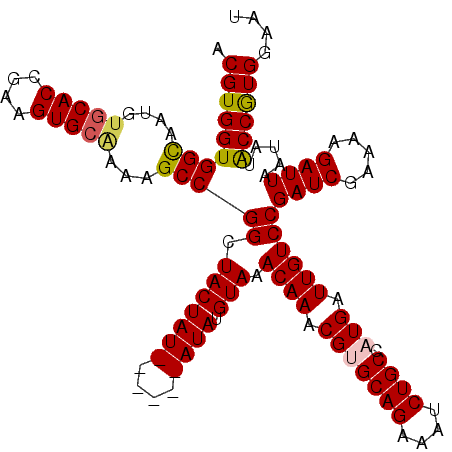
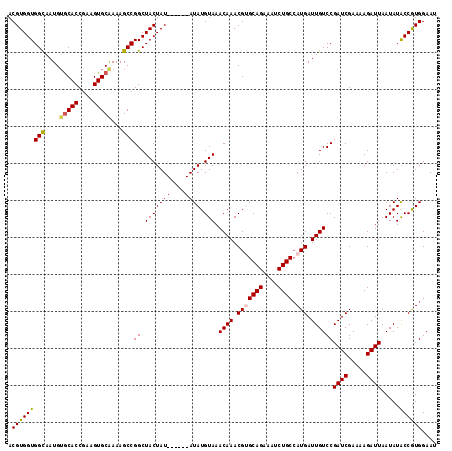
| Location | 6,410,297 – 6,410,407 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -25.72 |
| Energy contribution | -28.88 |
| Covariance contribution | 3.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6410297 110 + 22407834 CAGAUUUCUGCACGUUUGUUUACAUAU------AUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGAC ..((((((((.(((..(((((((....------...(....)(((...(((((....)))))....))).....)))))))..))))))))((((((....))))))..))).... ( -33.30) >DroSec_CAF1 38586 106 + 1 CAGAUUUCUGCGCGUUUGUUUACAUAU------AUAGUAGCCGGCUUUUGCACUUCGGUGUUCAUUACCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGA----GAC .....(((((.(((..(((((((....------..........((....)).....((((........))))..)))))))..))))))))((((((....))))))..----... ( -25.90) >DroSim_CAF1 39067 110 + 1 CAGAUUUCUGCGCGUUUGUUUACAUAU------AUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGAC ..((((((((.(((..(((((((....------...(....)(((...(((((....)))))....))).....)))))))..))))))))((((((....))))))..))).... ( -32.70) >DroEre_CAF1 37195 115 + 1 CAGAU-UCUGCACGUUUGUUUACAUAUACGUGUAUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGGCAGAAAGAUAUAUAAAUAUCUGAGUCAGAC ..(((-(((((((((.((....))...))))))).....(((((((.((((((....))))....(((......))))).)))))))....((((((....))))))))))).... ( -42.80) >DroYak_CAF1 42610 110 + 1 CAGAUUUCUGCACGUUUGUUUACAUAU------AUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAGGAUAUAUAAAUAUCUGAGUCAGAC ..((((((((.(((..(((((((....------...(....)(((....((((....)))).....))).....)))))))..))))))))((((((....))))))..))).... ( -31.70) >consensus CAGAUUUCUGCACGUUUGUUUACAUAU______AUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGAC .....(((((.(((..(((((((.((((....))))......(((....((((....)))).....))).....)))))))..))))))))((((((....))))))......... (-25.72 = -28.88 + 3.16)

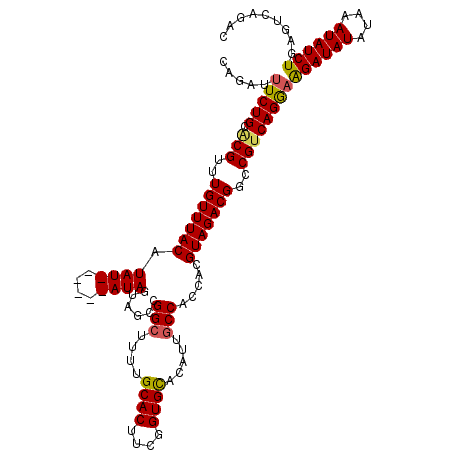
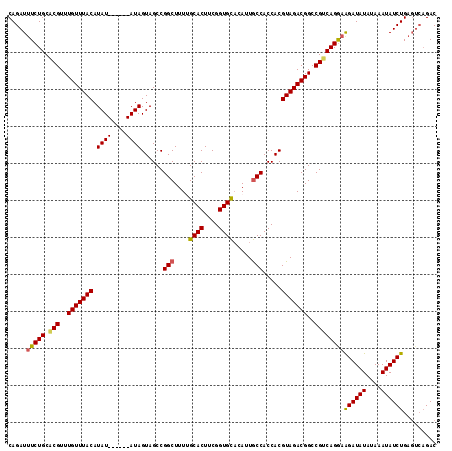
| Location | 6,410,327 – 6,410,447 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -33.52 |
| Energy contribution | -33.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6410327 120 + 22407834 GUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAACUGGAAACGCACAAUUUAAGUUGGAAA ....(((((((.(((((....)))))...(((..((.((....((((((((.((.((((((....))))))...)).))).).))))))))....(....))))......)))))))... ( -36.50) >DroSec_CAF1 38616 116 + 1 GUAGCCGGCUUUUGCACUUCGGUGUUCAUUACCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGA----GACUGUGCCGCGGGAGCUGGAAACGCACAAUUUAAGUGGGAAA ((..(((((((((((.....((((........))))........(((...(((..((((((....))))))..----..))).))))))))))))))..)).(((.......)))..... ( -38.70) >DroSim_CAF1 39097 119 + 1 GUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAGCUGGA-ACGCACAAUUUAAGUGGGAAA ....(((((((((((.....(((((.....).))))........(((((((.((.((((((....))))))...)).))).).)))))))))))))).-.(.(((.......))).)... ( -39.50) >DroEre_CAF1 37230 120 + 1 GUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGGCAGAAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAGUUGGAAACGCACAAUUUAAGAUGGAAG ...(((((((.((((((....))))....(((......))))).))))))).((.((((((....))))))...))(((.(((((.((....)).(....)))))).))).......... ( -40.50) >DroYak_CAF1 42640 120 + 1 GUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAGGAUAUAUAAAUAUCUGAGUCAGACUGUGCCACGGGAGUUGGAAACGCACAAUUUAAAUCGGAAA ((..((((((..(((((....))))...................(((((((.((.((((((....))))))...)).))).).)))..)..))))))..))................... ( -35.60) >consensus GUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAGCUGGAAACGCACAAUUUAAGUGGGAAA ((..(((((((((((.....(((((.....).))))........(((((((.((.((((((....))))))...)).))).).))))))))))))))..))................... (-33.52 = -33.44 + -0.08)

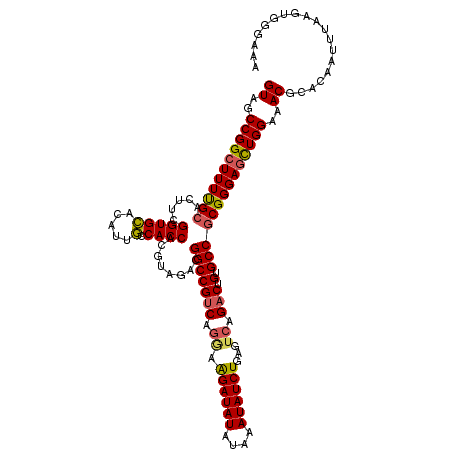
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:18 2006