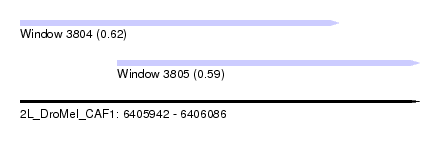
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,405,942 – 6,406,086 |
| Length | 144 |
| Max. P | 0.617304 |

| Location | 6,405,942 – 6,406,057 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.62 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6405942 115 + 22407834 CUCUGAGGGAUGAUCCAAAUCAAUGUCAG-----AGGAAAGGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCUGCCUACUUCCCCGAAACAUUUCCGGCU ((((((.(..((((....)))).).))))-----))....(((((..(((....)))......(..(((((((.((((((......)))))).)))).)))..).......))))).... ( -25.30) >DroSec_CAF1 34296 115 + 1 CUCUGAGGGAUGAUCCAAAUCAAUGUCAG-----AGGAAAGGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUACCCCGAAACAUUUCCGUGC ((((((.(..((((....)))).).))))-----))....(((((..(((....)))..........(((((((((((((......))))))))))).))...........))))).... ( -28.60) >DroSim_CAF1 34816 115 + 1 CUCUGAGGGAUGAUCCAAAUCAAUGUCAG-----AGGAAAGGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUACCCCGAAACAUUUCCGUGC ((((((.(..((((....)))).).))))-----))....(((((..(((....)))..........(((((((((((((......))))))))))).))...........))))).... ( -28.60) >DroEre_CAF1 32784 115 + 1 CUCUGAGGGAUGAUCCAAAUCAAUGUCAG-----AGCAAAGGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUGCUCCGAAACAUUUCCGGGC ((((((.(..((((....)))).).))))-----))....(((((..(((....))).((..((.((..(((((((((((......)))))))))))..))))..))....))))).... ( -29.60) >DroYak_CAF1 38109 120 + 1 CUCUGGGGGAUGAUCCAAAUCAAUGUCAGUGGAAAGGAAAGGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUUCUCCGAAACAUUUCCGGAC .((((((((.....)).....(((((...((((.(((...(....).......................(((((((((((......))))))))))).))).))))..))))))))))). ( -31.00) >consensus CUCUGAGGGAUGAUCCAAAUCAAUGUCAG_____AGGAAAGGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUACCCCGAAACAUUUCCGGGC ..((((.(..((((....)))).).))))......((((((....).....................(((((((((((((......))))))))))).))...........))))).... (-22.68 = -22.92 + 0.24)


| Location | 6,405,977 – 6,406,086 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -24.25 |
| Energy contribution | -24.61 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6405977 109 + 22407834 GGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCUGCCUACUUCCCCGAAACAUUUCCGGCUUGAA------AUAGGUGAGUUG--GGGGAAGCGACGU (....).................(((...((((.((((((......)))))).)))).((((((((((....)))((((((...------......))))))--))))))).))).. ( -30.10) >DroSec_CAF1 34331 109 + 1 GGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUACCCCGAAACAUUUCCGUGCUGAA------AUAGGUGAGUUG--AGGGAAGCGACGU (....).................(((.(((((((((((((......))))))))))).))..(((..(((.((.((........------...)).))))).--.)))....))).. ( -26.90) >DroSim_CAF1 34851 109 + 1 GGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUACCCCGAAACAUUUCCGUGCUGAA------AUAGGUGAGUUG--GGGGAAGCGACGU (....).................(((.(((((((((((((......))))))))))).)).((((((.(((((((......)))------))..))...)))--))).....))).. ( -30.50) >DroEre_CAF1 32819 109 + 1 GGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUGCUCCGAAACAUUUCCGGGCUUCU------AUAGGUGAGUUG--GGGGAAGCGACGU (....).................(((.(((((((((((((......))))))))))).))(((........(..((.(((((..------......))))).--))..))))))).. ( -28.50) >DroYak_CAF1 38149 117 + 1 GGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUUCUCCGAAACAUUUCCGGACUACUACGAGUAUAGGUGAGUUGUGGGGUAAGCGACGU (....).................(((...(((((((((((......))))))))))).((((((((.(((.((.((..(((......)))...)).))))).))))).))).))).. ( -30.50) >consensus GGAAACAUAAGAAAUUACUCAAAGUCAAGAGGUUGUUGAAACUAUGUUUAGCAGCCUACUACCCCGAAACAUUUCCGGGCUGAA______AUAGGUGAGUUG__GGGGAAGCGACGU (....).................(((...(((((((((((......))))))))))).((.((((..(((.((.((.................)).)))))...)))).)).))).. (-24.25 = -24.61 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:12 2006