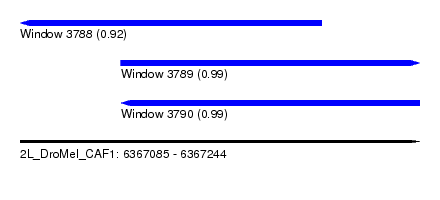
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,367,085 – 6,367,244 |
| Length | 159 |
| Max. P | 0.991980 |

| Location | 6,367,085 – 6,367,205 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -21.41 |
| Energy contribution | -22.09 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
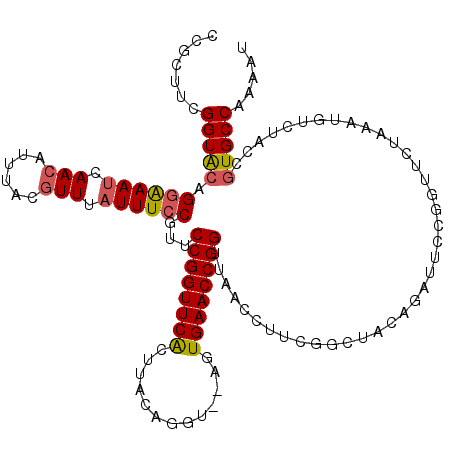
>2L_DroMel_CAF1 6367085 120 - 22407834 CCGCUUCGGUACAGGAAAUCAACAUUUACCUUUAUUUCCGUUCCGGUUCAGUUACAGUUACAGUGAACCGGUUAACCUUCGGCUACAGAUUCCGGUUCUAAAUGUUUACCGUGCCAAAAU .......(((((.((((.((...................(((((((((((.(.........).))))))))..)))(....).....))))))(((.(.....)...))))))))..... ( -27.30) >DroSec_CAF1 11959 118 - 1 CCGCUUCGGUGCAGGAAAUCAACAUUUACGUUUAUUUCCGUUCCGGUUCACUUACAGGU--AGUGAACCGGUUAACCUUCGGCUACAGAUUCCGGUUCUAAAUGUGUACCGUGCCAUAAU ..((..(((((((((((((.(((......))).))))))(((((((((((((.......--))))))))))..)))..((((.........)))).........))))))).))...... ( -38.60) >DroEre_CAF1 11841 116 - 1 CUGAUUCGGUAAAG-GAACUAACGCGUAUGUUUAUUUCCGUACCGGUUCGUUUACAAGU--ACUGAACCGGUUAAU-UUCGUUUACCGAUUUCGGUUCUAAGUUUCUUUCCUGCCAAAAU ((((.((((((((.-(((.((((..(((((........)))))((((((((........--)).))))))))))..-))).))))))))..))))......................... ( -30.90) >consensus CCGCUUCGGUACAGGAAAUCAACAUUUACGUUUAUUUCCGUUCCGGUUCACUUACAGGU__AGUGAACCGGUUAACCUUCGGCUACAGAUUCCGGUUCUAAAUGUCUACCGUGCCAAAAU .......(((((.((((((.(((......))).))))))...((((((((.............)))))))).......................................)))))..... (-21.41 = -22.09 + 0.67)

| Location | 6,367,125 – 6,367,244 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6367125 119 + 22407834 GAAGGUUAACCGGUUCACUGUAACUGUAACUGAACCGGAACGGAAAUAAAGGUAAAUGUUGAUUUCCUGUACCGAAGCGGUAGAACUGGUUAAGGGUUUAAACCUAAUUGAUUUCAUUU- ((((.((((((((((((((((....((......))(((.((((((((.((.(....).)).)))))).)).)))..))))).)))))))))))((((....))))......))))....- ( -32.70) >DroSec_CAF1 11999 116 + 1 GAAGGUUAACCGGUUCACU--ACCUGUAAGUGAACCGGAACGGAAAUAAACGUAAAUGUUGAUUUCCUGCACCGAAGCGGUUAAGCUUGC-GAGGUUUCACGCCUAAUUGAUUUAAUUU- ..((((...((((((((((--.......))))))))))...((((((.((((....)))).)))))).((..((((((......)))).)-)..)).....))))..............- ( -35.50) >DroEre_CAF1 11881 113 + 1 GAA-AUUAACCGGUUCAGU--ACUUGUAAACGAACCGGUACGGAAAUAAACAUACGCGUUAGUUC-CUUUACCGAAUCAGUAAACCGG---ACCGGUUCGAACCCAUUUAUUUACAUUUU ...-.......(((((.((--........))((((((((..((((...(((......)))..)))-)....(((...........)))---)))))))))))))................ ( -27.90) >consensus GAAGGUUAACCGGUUCACU__ACCUGUAAAUGAACCGGAACGGAAAUAAACGUAAAUGUUGAUUUCCUGUACCGAAGCGGUAAAACUGG__AAGGGUUCAAACCUAAUUGAUUUCAUUU_ .........((((((((.............))))))))...((((((.(((......))).))))))...........(((.(((((......)))))...)))................ (-17.94 = -18.62 + 0.68)


| Location | 6,367,125 – 6,367,244 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6367125 119 - 22407834 -AAAUGAAAUCAAUUAGGUUUAAACCCUUAACCAGUUCUACCGCUUCGGUACAGGAAAUCAACAUUUACCUUUAUUUCCGUUCCGGUUCAGUUACAGUUACAGUGAACCGGUUAACCUUC -..............(((((..................(((((...)))))..((((((.((........)).))))))...((((((((.(.........).))))))))..))))).. ( -22.40) >DroSec_CAF1 11999 116 - 1 -AAAUUAAAUCAAUUAGGCGUGAAACCUC-GCAAGCUUAACCGCUUCGGUGCAGGAAAUCAACAUUUACGUUUAUUUCCGUUCCGGUUCACUUACAGGU--AGUGAACCGGUUAACCUUC -............((((((((((....))-))..)))))).(((....)))..((((((.(((......))).))))))(((((((((((((.......--))))))))))..))).... ( -35.50) >DroEre_CAF1 11881 113 - 1 AAAAUGUAAAUAAAUGGGUUCGAACCGGU---CCGGUUUACUGAUUCGGUAAAG-GAACUAACGCGUAUGUUUAUUUCCGUACCGGUUCGUUUACAAGU--ACUGAACCGGUUAAU-UUC .......((((.(((.(((((((((((((---.((((((((((...))))))).-((((..........))))....))).))))))))((........--)).))))).))).))-)). ( -32.60) >consensus _AAAUGAAAUCAAUUAGGUUUGAACCCUU__CCAGCUUUACCGCUUCGGUACAGGAAAUCAACAUUUACGUUUAUUUCCGUUCCGGUUCACUUACAGGU__AGUGAACCGGUUAACCUUC ......................................(((((...)))))..((((((.(((......))).))))))...((((((((.............))))))))......... (-17.28 = -17.95 + 0.67)

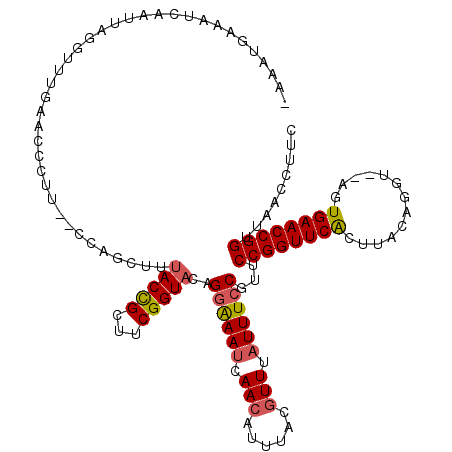
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:57 2006