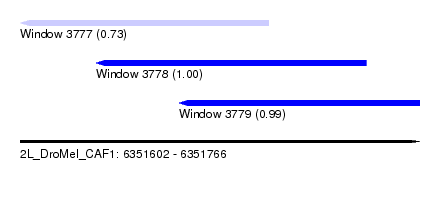
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,351,602 – 6,351,766 |
| Length | 164 |
| Max. P | 0.998099 |

| Location | 6,351,602 – 6,351,704 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6351602 102 - 22407834 U-GACGGGCGGUGUUCCAGCAGCGAAGCACCGCCUGUUGUCAAUCCGAACUAUC-AGCCGAAUCGAAUCUCAA--------UCUCUCCAUAGAGCACCU---AUGACGUGGUC---GA .-(((((((((((((.(......).))))))))))))).......(((.((((.-....((........))..--------......(((((.....))---)))..))))))---). ( -29.80) >DroSec_CAF1 3345 102 - 1 U-GACGGGCGGUGUUCCAGCAGCGGAGCACCGCCUGUUGUCAAACCGAACUAUC-AACCGAAUCGAAUCUCAA--------UCUCUCAAUAGAGCACCU---AUGACGUGGUC---GA .-(((((((((((((((......)))))))))))))))..............((-.((((..(((........--------.((((....)))).....---.)))..)))).---)) ( -35.36) >DroSim_CAF1 2036 102 - 1 U-GACGGGCGGUGUUCCAGCAGCGGAGCACCGCCUGUUGUCAAACCGAACUAUC-AACCGAAUCGAAUCUCAA--------UCUCUCAAUAGAGCACCU---AUGACGUGGCC---GA .-(((((((((((((((......)))))))))))))))((((...(((....((-....)).)))........--------.((((....)))).....---.))))......---.. ( -34.70) >DroEre_CAF1 2047 87 - 1 U-GACGGGUGUUGUUCCAGCAGCGAAGCACCGCCUG-------------CUAUC-AAGC--AUCGAAUCUCAA--------UCUCUGCACAGAGCACCU---GUGACGUGGUC---GA .-.(((((((((.....(((((((......)).)))-------------))...-..((--(..((.......--------))..)))....)))))))---))(((...)))---.. ( -24.30) >DroYak_CAF1 2333 103 - 1 U-GACGGGCGGUGUUCCAGCAGUGAAGCACCGCCUGUUGUUUAACUCAACUAUC-AAGC--AACGAAUCUCAA--------UCUUUCAAUAGAGCACCUACUAUGACGUGGUG---GA .-(((((((((((((.((....)).)))))))))))))...........(((((-(..(--(...........--------(((......)))..........))...)))))---). ( -29.60) >DroAna_CAF1 2272 107 - 1 UGGAAAGGCGGUGUUCCAGCGCUGGAGCACCGCCUGUC-------CGA-CUGCCGCGGCUACUCGCAUAUCAACAAUCAAGCCCCUCGAGAGAGUUCCA---AUGACGUGGGCCCCGA .(((.(((((((((((((....))))))))))))).))-------)..-...((((((((...................)))).(((....))).....---.....))))....... ( -44.31) >consensus U_GACGGGCGGUGUUCCAGCAGCGAAGCACCGCCUGUUGUCAAACCGAACUAUC_AACCGAAUCGAAUCUCAA________UCUCUCAAUAGAGCACCU___AUGACGUGGUC___GA ..(((((((((((((.(......).)))))))))))))............................................((((....)))).(((...........)))...... (-20.61 = -20.87 + 0.25)

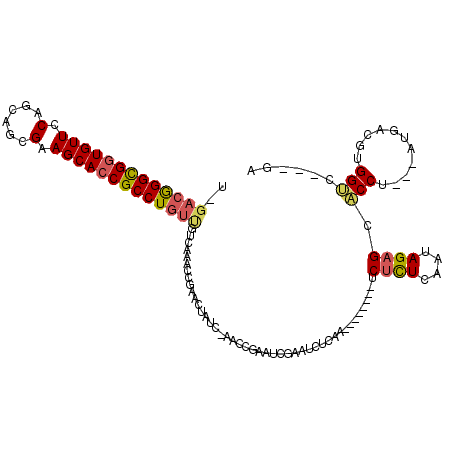
| Location | 6,351,633 – 6,351,744 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.94 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6351633 111 - 22407834 UUGAUAACACCCGGAGCAAAUACCACGUCUGUUGACCCACU-GACGGGCGGUGUUCCAGCAGCGAAGCACCGCCUGUUGUCAAUCCGAACUAUC-AGCCGAAUCGAAUCUCAA----- ((((((.....((((...........(((....))).....-(((((((((((((.(......).))))))))))))).....))))...))))-))................----- ( -31.90) >DroSec_CAF1 3376 89 - 1 UUGAUAACAACCG----------------------CCCACU-GACGGGCGGUGUUCCAGCAGCGGAGCACCGCCUGUUGUCAAACCGAACUAUC-AACCGAAUCGAAUCUCAA----- (((((((((...(----------------------(((...-...))))((((((((......))))))))...)))))))))...........-..................----- ( -31.70) >DroSim_CAF1 2067 111 - 1 UUGAUAACACCGGGAGCAAAUACCCGGUCUGUUGACCCACU-GACGGGCGGUGUUCCAGCAGCGGAGCACCGCCUGUUGUCAAACCGAACUAUC-AACCGAAUCGAAUCUCAA----- ((((((..((((((........))))))...(((((.....-(((((((((((((((......)))))))))))))))))))).......))))-))................----- ( -45.70) >DroEre_CAF1 2078 96 - 1 UUGAUAACACCCGGAGCAAAUACCAGGUGUGUUGACCCACU-GACGGGUGUUGUUCCAGCAGCGAAGCACCGCCUG-------------CUAUC-AAGC--AUCGAAUCUCAA----- ((((((......(((((((.((((.((((.(....).))))-....))))))))))).((((((......)).)))-------------)))))-))..--............----- ( -29.30) >DroYak_CAF1 2367 109 - 1 UUGAUAACACCGAGAACAAAUACGAGGUCUGUUGACCCACU-GACGGGCGGUGUUCCAGCAGUGAAGCACCGCCUGUUGUUUAACUCAACUAUC-AAGC--AACGAAUCUCAA----- ((((((.....(((.........(.((((....)))))...-(((((((((((((.((....)).)))))))))))))......)))...))))-))..--............----- ( -34.20) >DroAna_CAF1 2309 102 - 1 UUGAUAAUACGAAAAACAAAUAUUAG--------CCCCCCUGGAAAGGCGGUGUUCCAGCGCUGGAGCACCGCCUGUC-------CGA-CUGCCGCGGCUACUCGCAUAUCAACAAUC ((((((....................--------......((((.(((((((((((((....))))))))))))).))-------)).-.....((((....)))).))))))..... ( -38.80) >consensus UUGAUAACACCCGGAGCAAAUACCAGGUCUGUUGACCCACU_GACGGGCGGUGUUCCAGCAGCGAAGCACCGCCUGUUGUCAAACCGAACUAUC_AACCGAAUCGAAUCUCAA_____ ..........................................(((((((((((((.(......).)))))))))))))........................................ (-19.44 = -19.72 + 0.28)


| Location | 6,351,667 – 6,351,766 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -18.97 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6351667 99 - 22407834 GAUACGGAUGCCUCAAGCAGAGUUGAUAACACCCGGAGCAAAUACCACGUCUGUUGACCCACU-GACGGGCGGUGUUCCAGCAGCGAAGCACCGCCUGUU ...(((((((.(((.....)))............((........)).))))))).........-(((((((((((((.(......).))))))))))))) ( -30.60) >DroSec_CAF1 3410 77 - 1 GAUACGGAUGCCUCAAGCAGAGUUGAUAACAACCG----------------------CCCACU-GACGGGCGGUGUUCCAGCAGCGGAGCACCGCCUGUU .....((.(((.....)))(.((((....))))).----------------------.))...-(((((((((((((((......))))))))))))))) ( -34.00) >DroSim_CAF1 2101 99 - 1 GAUACGGAUGCCUCAAGCAGAGUUGAUAACACCGGGAGCAAAUACCCGGUCUGUUGACCCACU-GACGGGCGGUGUUCCAGCAGCGGAGCACCGCCUGUU .....((.(((.....)))..((..(((..((((((........)))))).)))..))))...-(((((((((((((((......))))))))))))))) ( -46.70) >DroEre_CAF1 2099 97 - 1 GAUGCGGACACCUCAAGCAGAGUUGAUAACACCCGGAGCAAAUACCAGGUGUGUUGACCCACU-GACGGGUGUUGUUCCAGCAGCGAAGCACCGCCUG-- ...((((((((((....(((.((..(((.((((.((........)).)))))))..))...))-)..)))))).......((......)).))))...-- ( -31.00) >DroYak_CAF1 2399 99 - 1 AAUCCGGAUACCUCCAGCACAGUUGAUAACACCGAGAACAAAUACGAGGUCUGUUGACCCACU-GACGGGCGGUGUUCCAGCAGUGAAGCACCGCCUGUU ..(((((.......((((...))))......))).))........(.((((....)))))...-(((((((((((((.((....)).))))))))))))) ( -32.32) >DroAna_CAF1 2341 76 - 1 ACU----------------AGGUUGAUAAUACGAAAAACAAAUAUUAG--------CCCCCCUGGAAAGGCGGUGUUCCAGCGCUGGAGCACCGCCUGUC ...----------------.((((((((..............))))))--------))......((.(((((((((((((....))))))))))))).)) ( -31.54) >consensus GAUACGGAUGCCUCAAGCAGAGUUGAUAACACCCGGAGCAAAUACCAGGUCUGUUGACCCACU_GACGGGCGGUGUUCCAGCAGCGAAGCACCGCCUGUU ................................................................(((((((((((((.(......).))))))))))))) (-18.97 = -19.25 + 0.28)

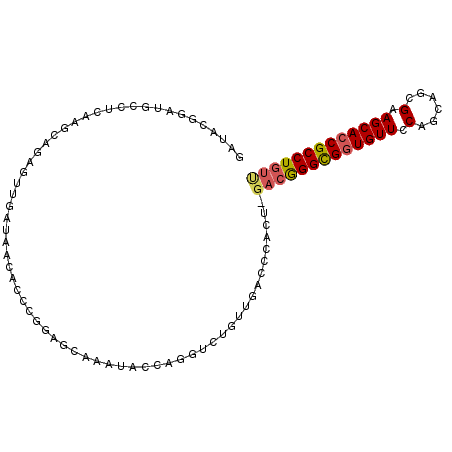
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:47 2006