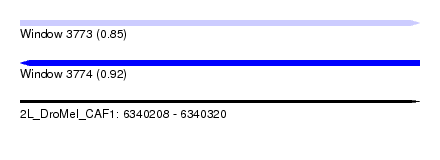
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,340,208 – 6,340,320 |
| Length | 112 |
| Max. P | 0.918094 |

| Location | 6,340,208 – 6,340,320 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -12.00 |
| Energy contribution | -14.88 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.852737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6340208 112 + 22407834 UUAUGACACCCCAUUAUAGGGUUACCUACCUAUUAAAGC--------UAGACUAUUAGUUCUUCCAAUUAAAGAACCAUUAAAAGGGCAGGUUCCGUAAUGGGAUUACAUUUAAUGGACU .........((((((((.(((..((((.(((.((((((.--------....))....((((((.......))))))..)))).)))..)))))))))))))))................. ( -24.30) >DroSec_CAF1 31280 120 + 1 UUAUGAGACCCCAUUAUGGGGUUACCUACCUAUUAAAGACUAUUAUAAAGACUAUUAGUUCUUCCACUUAAAGAACCAUUAAAAGGGUAGGUUCCGUAUUGGGGUUACAUUUAAUGGAUU ..(((.(((((((.((((((...((((((((.....((.((.......)).))....((((((.......))))))........)))))))))))))).))))))).))).......... ( -35.60) >DroSim_CAF1 33243 112 + 1 UCAUGACACCCCAUUAUGGGGUUACCUACCUAUUAAAGC--------UAGACUAUUAGUUCUUCCACUUAAAGAACCAUUAAAAGGGUAGGUUCCGUAUUGGGGUUACAUUUAAUGGAUU ..(((..((((((.((((((...((((((((.....((.--------....))....((((((.......))))))........)))))))))))))).))))))..))).......... ( -32.40) >DroEre_CAF1 31942 109 + 1 UUAUGACACCCCAUUAUGGGGUUACCUACCUAGUAAAGC--------UAGACUUCUAGAUCUUCCACUCAAAGUACCGAUAAAAAGCUAAGGUCAGGAUUAGGGUUGC---UGGGUUGGU .......(((((.....))))).(((.(((((((((..(--------(((..((((.((((((........(((...........)))))))))))))))))..))))---))))).))) ( -31.40) >DroYak_CAF1 31378 111 + 1 CUAUGAC-CCCCAUUAUGGGGUUACCUACCUAUUAACGC--------UAGACCCUUAGAUCUGCCACACAAAGAACCAUUAAAAGGGUAAGGUCGGUUUUGGGGUUACAUUUAAUUAAUU ...((((-(((......)))))))...............--------..((((((.(((((.(((.(.....).(((........)))..))).))))).)))))).............. ( -27.20) >consensus UUAUGACACCCCAUUAUGGGGUUACCUACCUAUUAAAGC________UAGACUAUUAGUUCUUCCACUUAAAGAACCAUUAAAAGGGUAGGUUCCGUAUUGGGGUUACAUUUAAUGGAUU .......((((((.(((.(((...(((((((................(((....)))((((((.......))))))........))))))).)))))).))))))............... (-12.00 = -14.88 + 2.88)

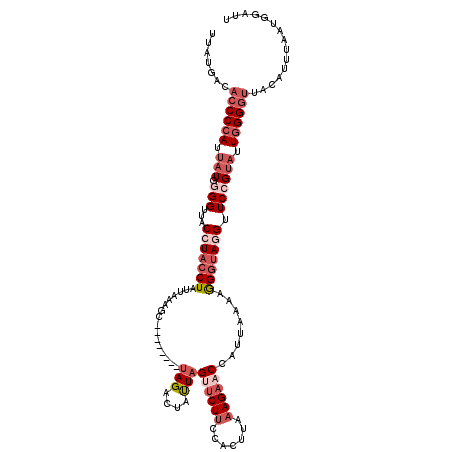
| Location | 6,340,208 – 6,340,320 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -17.12 |
| Energy contribution | -19.32 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6340208 112 - 22407834 AGUCCAUUAAAUGUAAUCCCAUUACGGAACCUGCCCUUUUAAUGGUUCUUUAAUUGGAAGAACUAAUAGUCUA--------GCUUUAAUAGGUAGGUAACCCUAUAAUGGGGUGUCAUAA ..........(((..(((((((((..(.(((((((...(((((((((((((.....)))))))))..((....--------.))))))..)))))))....)..)))))))))..))).. ( -31.50) >DroSec_CAF1 31280 120 - 1 AAUCCAUUAAAUGUAACCCCAAUACGGAACCUACCCUUUUAAUGGUUCUUUAAGUGGAAGAACUAAUAGUCUUUAUAAUAGUCUUUAAUAGGUAGGUAACCCCAUAAUGGGGUCUCAUAA ..........(((..((((((.((.((.(((((((...(((((((((((((.....)))))))))..((.((.......)).))))))..)))))))....)).)).))))))..))).. ( -34.00) >DroSim_CAF1 33243 112 - 1 AAUCCAUUAAAUGUAACCCCAAUACGGAACCUACCCUUUUAAUGGUUCUUUAAGUGGAAGAACUAAUAGUCUA--------GCUUUAAUAGGUAGGUAACCCCAUAAUGGGGUGUCAUGA ..........(((..((((((.((.((.(((((((...(((((((((((((.....)))))))))..((....--------.))))))..)))))))....)).)).))))))..))).. ( -31.70) >DroEre_CAF1 31942 109 - 1 ACCAACCCA---GCAACCCUAAUCCUGACCUUAGCUUUUUAUCGGUACUUUGAGUGGAAGAUCUAGAAGUCUA--------GCUUUACUAGGUAGGUAACCCCAUAAUGGGGUGUCAUAA (((.(((.(---(...((((.......(((.............)))......)).)).(((.((((....)))--------).))).)).))).))).(((((.....)))))....... ( -23.64) >DroYak_CAF1 31378 111 - 1 AAUUAAUUAAAUGUAACCCCAAAACCGACCUUACCCUUUUAAUGGUUCUUUGUGUGGCAGAUCUAAGGGUCUA--------GCGUUAAUAGGUAGGUAACCCCAUAAUGGGG-GUCAUAG ..........................(((((((((...(((((((((........)))(((((....))))).--------.))))))..))))))...((((.....))))-))).... ( -24.80) >consensus AAUCCAUUAAAUGUAACCCCAAUACGGAACCUACCCUUUUAAUGGUUCUUUAAGUGGAAGAACUAAUAGUCUA________GCUUUAAUAGGUAGGUAACCCCAUAAUGGGGUGUCAUAA ..........(((..((((((.((.((.(((((((.......(((((((((.....)))))))))...(.............).......)))))))....)).)).))))))..))).. (-17.12 = -19.32 + 2.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:42 2006