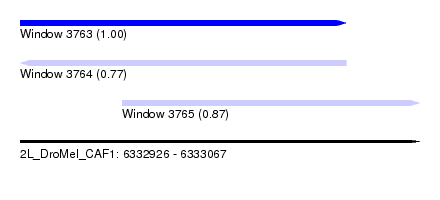
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,332,926 – 6,333,067 |
| Length | 141 |
| Max. P | 0.997987 |

| Location | 6,332,926 – 6,333,041 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 96.61 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6332926 115 + 22407834 UACACAUGUAUAAAUAGUUCGGCUUUAAACCUAAGCCCAAUUGUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAU .......((((((((((((.(((((.......))))).)))))))(((((.((((.(((((...(((((............)))))....))))).))))..)))))...))))) ( -28.90) >DroSec_CAF1 24047 115 + 1 UACACAUGUAUAAAUAGUUCGGCUUUAAACCUAAGCCCAAUUGUUGGGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAU .......(((((((.(((.(((...........(((((........)))))((((((.(((...(((((............))))).)))))))))....)))..)))))))))) ( -27.70) >DroSim_CAF1 25983 115 + 1 UACACAUGUAUAAAUAGUUCGGCUUUAAACCUAAGCCCAAUUGUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAU .......((((((((((((.(((((.......))))).)))))))(((((.((((.(((((...(((((............)))))....))))).))))..)))))...))))) ( -28.90) >DroEre_CAF1 24503 115 + 1 UACACAUGUAUAAAUACAUCGGCUUUAAACCUAAGCCCAAUUUUGGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUGCGCUAUUUUUAUAU .......(((((((......(((((.......))))).......(((((((((((.(((((...(((((............)))))....))))).))))).))))))))))))) ( -25.30) >DroYak_CAF1 24001 115 + 1 UACACAUGUAUAAAUAGUUCGGCUUUAUACCUAAGCCCAAUUUUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUUCGCUAUUUUUAUAU .....(((((.(((((((..(((((.......)))))........((((..((((((.(((...(((((............))))).)))))))))))))..))))))).))))) ( -25.40) >consensus UACACAUGUAUAAAUAGUUCGGCUUUAAACCUAAGCCCAAUUGUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAU .......((((((((((((.(((((.......))))).)))))).(((((.((((.(((((...(((((............)))))....))))).))))..)))))..)))))) (-24.38 = -25.14 + 0.76)

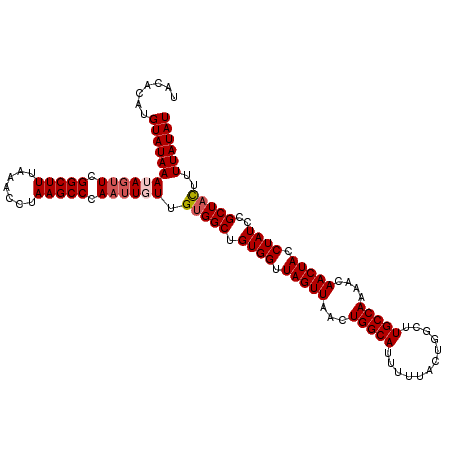
| Location | 6,332,926 – 6,333,041 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.61 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6332926 115 - 22407834 AUAUAAAAGUAGCGGAUAGGUAGUUGUUUUGGCAAGCCAGUAAAAAUGCCAGUUAACUAACCACAGCCACAACAAUUGGGCUUAGGUUUAAAGCCGAACUAUUUAUACAUGUGUA .(((((((((..(((...((((((((..((((((............)))))).))))).)))..((((.((.....))))))...........))).))).))))))........ ( -25.70) >DroSec_CAF1 24047 115 - 1 AUAUAAAAGUAGCGGAUAGGUAGUUGUUUUGGCAAGCCAGUAAAAAUGCCAGUUAACUAACCACAGCCCCAACAAUUGGGCUUAGGUUUAAAGCCGAACUAUUUAUACAUGUGUA .(((((((((..(((...((((((((..((((((............)))))).))))).)))..(((((........)))))...........))).))).))))))........ ( -27.90) >DroSim_CAF1 25983 115 - 1 AUAUAAAAGUAGCGGAUAGGUAGUUGUUUUGGCAAGCCAGUAAAAAUGCCAGUUAACUAACCACAGCCACAACAAUUGGGCUUAGGUUUAAAGCCGAACUAUUUAUACAUGUGUA .(((((((((..(((...((((((((..((((((............)))))).))))).)))..((((.((.....))))))...........))).))).))))))........ ( -25.70) >DroEre_CAF1 24503 115 - 1 AUAUAAAAAUAGCGCAUAGGUAGUUGUUUUGGCAAGCCAGUAAAAAUGCCAGUUAACUAACCACAGCCACCAAAAUUGGGCUUAGGUUUAAAGCCGAUGUAUUUAUACAUGUGUA ...........((((((.((((((((..((((((............)))))).))))).)))((((((..........)))...(((.....)))..)))........)))))). ( -26.00) >DroYak_CAF1 24001 115 - 1 AUAUAAAAAUAGCGAAUAGGUAGUUGUUUUGGCAAGCCAGUAAAAAUGCCAGUUAACUAACCACAGCCACAAAAAUUGGGCUUAGGUAUAAAGCCGAACUAUUUAUACAUGUGUA ......((((((......((((((((..((((((............)))))).))))).)))..((((.((.....))))))..(((.....)))...))))))........... ( -23.80) >consensus AUAUAAAAGUAGCGGAUAGGUAGUUGUUUUGGCAAGCCAGUAAAAAUGCCAGUUAACUAACCACAGCCACAACAAUUGGGCUUAGGUUUAAAGCCGAACUAUUUAUACAUGUGUA ......((((((......((((((((..((((((............)))))).))))).)))..((((.((.....))))))..(((.....)))...))))))........... (-22.54 = -22.70 + 0.16)

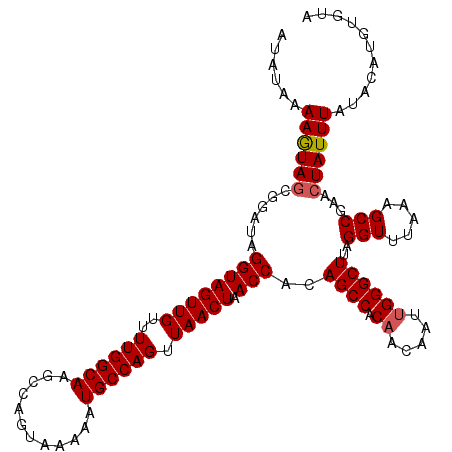
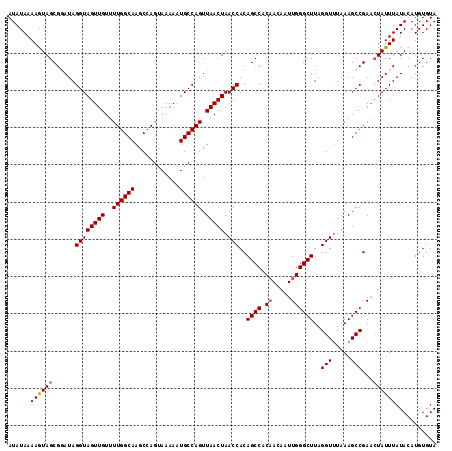
| Location | 6,332,962 – 6,333,067 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.69 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
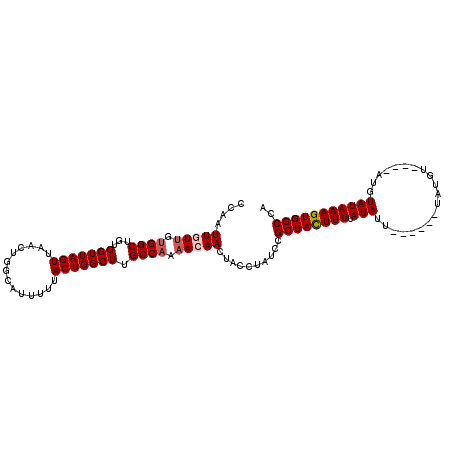
>2L_DroMel_CAF1 6332962 105 + 22407834 CCAAUUGUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAUU------UAUGUAUGUAUCUAUAAAGUGGCCA ....(((((.((((...(((((((..............))))))).)))).)))))..........((((((((.(((..------(((....)))..))))))))))).. ( -25.84) >DroSec_CAF1 24083 101 + 1 CCAAUUGUUGGGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAUU------UAUGU----AUUUAUAAAGUGGCCA .........(((...((((((.(((...(((((............))))).)))))))))...)))((((((((.((((.------...))----))....)))))))).. ( -25.30) >DroSim_CAF1 26019 101 + 1 CCAAUUGUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAUU------UAUGU----AUUUAUAAAGUGGCCA ....(((((.((((...(((((((..............))))))).)))).)))))..........((((((((.((((.------...))----))....)))))))).. ( -25.24) >DroEre_CAF1 24539 107 + 1 CCAAUUUUGGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUGCGCUAUUUUUAUAUACGAGUAUAUGU----AUGUAUAAAGUGGCCA ........(((((.(((.(((((....)))))........(((....)))..))).)))))..((.((((((((((((((((......)))----))))).)))))))))) ( -28.50) >DroYak_CAF1 24037 97 + 1 CCAAUUUUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUUCGCUAUUUUUAUAUA----------U----AUGUAUAAAGUGGCCA ..........(((((((((((.(((...(((((............))))).)))))))))...........((((((((.----------.----..)))))))).))))) ( -20.90) >consensus CCAAUUGUUGUGGCUGUGGUUAGUUAACUGGCAUUUUUACUGGCUUGCCAAAACAACUACCUAUCCGCUACUUUUAUAUU______UAUGU____AUGUAUAAAGUGGCCA ....(((((.((((...(((((((..............))))))).)))).)))))..........((((((((.(((....................))))))))))).. (-19.13 = -19.69 + 0.56)

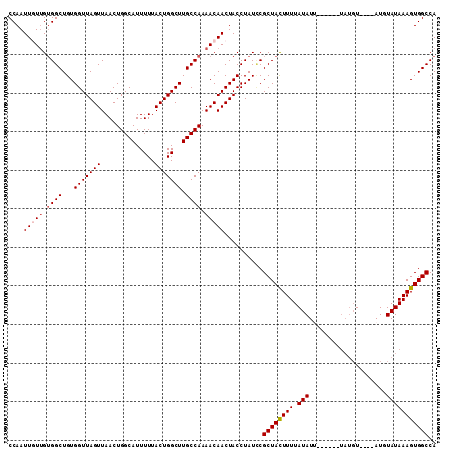
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:34 2006