
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,325,520 – 6,325,632 |
| Length | 112 |
| Max. P | 0.975275 |

| Location | 6,325,520 – 6,325,632 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -29.10 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
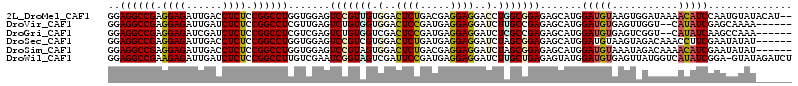
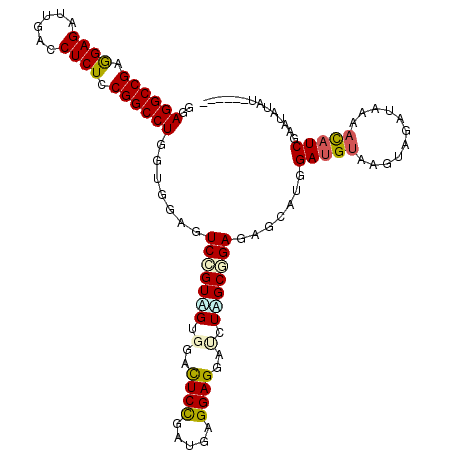
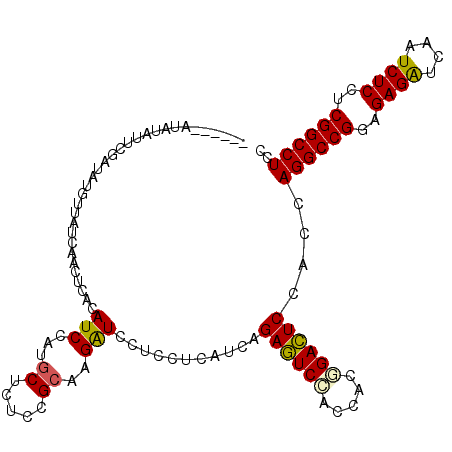
>2L_DroMel_CAF1 6325520 112 + 22407834 GGAGGCCGAGGAGAUUGACCUCUCCGGCCUGGUGGAGUCCGUUGUGGACUCUGACGAGGAGGACCUGGCGGAGAGCAUGGAUGUAAGUGGAUAAAACAUCCAAUGUAUACAU-- ..((((((..(((......)))..)))))).(((...(((((((.(..((((.....))))..).)))))))..((((((((((...........)))))).)))).)))..-- ( -42.30) >DroVir_CAF1 34921 106 + 1 GGAGGCCGAGGAGAUUGAUCUCUCCGGCCUCGUUGAGUCUGUGGUGGACUCCGAUGAGGAGGAUCUUGCCGAGAGCAUGGAUGUGAGUUGGU--CAUAUCGAGCAAAA------ .(((((((..((((....))))..)))))))......(((.(((..(.((((.....))))....)..))))))((.(.(((((((.....)--)))))).)))....------ ( -44.00) >DroGri_CAF1 8919 106 + 1 GGAGGCCGAGGAGAUCGAUCUCUCCGGCCUCGUCGAGUCUGUGGUCGACUCCGAUGAGGAGGAUCUCGCCGAGAGCAUGGAUGUGAGUCGGU--CAUAUCAAGCCAAA------ .(((((((..((((....))))..)))))))........((((..((((((((.((((......)))).))...(((....)))))))))..--))))..........------ ( -42.50) >DroSec_CAF1 16640 108 + 1 GGAGGCCGAGGAGAUUGACCUCUCCGGCCUGGUGGAGUCCGUCGUGGACUCUGAUGAGGAGGAUCUAGCGGAGAGCAUGGAUGUAAGUAGACAAACCUUCGAAUAUAU------ ..((((((..(((......)))..))))))...(((((((.....))))))).....((((((((((((.....)).)))))((......))...)))))........------ ( -39.40) >DroSim_CAF1 16829 108 + 1 GGAGGCCGAGGAGAUUGACCUCUCCGGCCUGGUGGAGUCCGUAGUGGACUCUGACGAGGAGGAUCUAGCGGAGAGCAUGGAUGUAAAUAGACAAAACAUCGAAUAUAU------ ..((((((..(((......)))..))))))((((((((((.....)))))).........(.(((((((.....)).))))).)............))))........------ ( -36.10) >DroWil_CAF1 57563 113 + 1 GGAGGCCGAAGAGAUUGAUCUCUCCGGCCUUGUCGAAUCGGUAGUCGAUUCCGAUGAGGAGGAUCUUGCUGAGAGUAUGGAUGUGAGUUAUGGUCAUAUCGGA-GUAUAGAUCU .(((((((.(((((....))))).)))))))(((((((((.....))))).)))).....((((((((((.......(.(((((((.......))))))).))-))).)))))) ( -39.31) >consensus GGAGGCCGAGGAGAUUGACCUCUCCGGCCUGGUGGAGUCCGUAGUGGACUCCGAUGAGGAGGAUCUAGCGGAGAGCAUGGAUGUAAGUAGAUAAAACAUCGAAUAUAU______ ..((((((.((((......)))).)))))).......(((((((.(..((((.....))))..).))))))).......(((((...........))))).............. (-29.10 = -29.13 + 0.03)
| Location | 6,325,520 – 6,325,632 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
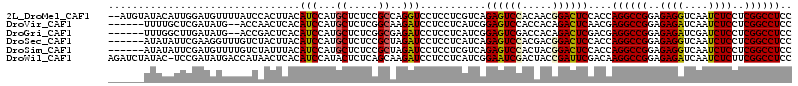
>2L_DroMel_CAF1 6325520 112 - 22407834 --AUGUAUACAUUGGAUGUUUUAUCCACUUACAUCCAUGCUCUCCGCCAGGUCCUCCUCGUCAGAGUCCACAACGGACUCCACCAGGCCGGAGAGGUCAAUCUCCUCGGCCUCC --..........(((((((...........)))))))........((.(((....))).))..((((((.....))))))....((((((..((((....))))..)))))).. ( -36.20) >DroVir_CAF1 34921 106 - 1 ------UUUUGCUCGAUAUG--ACCAACUCACAUCCAUGCUCUCGGCAAGAUCCUCCUCAUCGGAGUCCACCACAGACUCAACGAGGCCGGAGAGAUCAAUCUCCUCGGCCUCC ------((((((.(((((((--.............))))...)))))))))............(((((.......)))))...(((((((..((((....))))..))))))). ( -34.42) >DroGri_CAF1 8919 106 - 1 ------UUUGGCUUGAUAUG--ACCGACUCACAUCCAUGCUCUCGGCGAGAUCCUCCUCAUCGGAGUCGACCACAGACUCGACGAGGCCGGAGAGAUCGAUCUCCUCGGCCUCC ------...(((..(((.((--(.....))).)))...)))..((..(((......)))..))(((((.......)))))...(((((((..((((....))))..))))))). ( -37.80) >DroSec_CAF1 16640 108 - 1 ------AUAUAUUCGAAGGUUUGUCUACUUACAUCCAUGCUCUCCGCUAGAUCCUCCUCAUCAGAGUCCACGACGGACUCCACCAGGCCGGAGAGGUCAAUCUCCUCGGCCUCC ------........((.((...(((((...........((.....)))))))...))))....((((((.....))))))....((((((..((((....))))..)))))).. ( -30.80) >DroSim_CAF1 16829 108 - 1 ------AUAUAUUCGAUGUUUUGUCUAUUUACAUCCAUGCUCUCCGCUAGAUCCUCCUCGUCAGAGUCCACUACGGACUCCACCAGGCCGGAGAGGUCAAUCUCCUCGGCCUCC ------........((((....(((((...........((.....)))))))......)))).((((((.....))))))....((((((..((((....))))..)))))).. ( -30.50) >DroWil_CAF1 57563 113 - 1 AGAUCUAUAC-UCCGAUAUGACCAUAACUCACAUCCAUACUCUCAGCAAGAUCCUCCUCAUCGGAAUCGACUACCGAUUCGACAAGGCCGGAGAGAUCAAUCUCUUCGGCCUCC .(((((...(-(..((((((...............)))).))..))..)))))..........((((((.....))))))....((((((((((((....)))))))))))).. ( -32.46) >consensus ______AUAUAUUCGAUAUGUUAUCAACUCACAUCCAUGCUCUCCGCAAGAUCCUCCUCAUCAGAGUCCACCACGGACUCCACCAGGCCGGAGAGAUCAAUCUCCUCGGCCUCC ................................(((...((.....))..)))...........((((((.....))))))....((((((..((((....))))..)))))).. (-24.43 = -24.10 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:27 2006