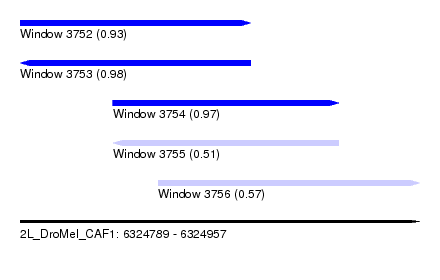
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,324,789 – 6,324,957 |
| Length | 168 |
| Max. P | 0.984711 |

| Location | 6,324,789 – 6,324,886 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6324789 97 + 22407834 GUGUGCAUCGCAAUCAAAGCUCCGCAGAUGCCACUACAUCCG--CG-------------------AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA-CGCUAUACAAUGGCUAAUCU (((.(((((((............)).)))))))).......(--((-------------------.(((((((((..............)))))))))-)))................. ( -29.14) >DroSec_CAF1 15893 97 + 1 GUGUGCAUCGCAAUCAAAGCUUCGCAGAUGCCACUACAUCCG--CG-------------------AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA-CGCAAUACAAUGGCUAAUCU (((.(((((((............)).)))))))).......(--((-------------------.(((((((((..............)))))))))-)))................. ( -29.64) >DroSim_CAF1 16094 97 + 1 GUGUGCAUCGCAAUCAAAGCUCCGCAGAUGCCACUACAUCCG--CG-------------------AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA-CGCAAUACAAUGGCUAAUCU (((.(((((((............)).)))))))).......(--((-------------------.(((((((((..............)))))))))-)))................. ( -29.64) >DroEre_CAF1 16205 115 + 1 GUGUGCAUCGCAAUCAAAGCUCUGCAGAUGCCACUACAUCAG--CGAUUUCUAUGAGCC--GCCGAUUCGGGCCACCCUUUCUUCAAAUUGCCCCAAAACGCAAUACAAUGGCUAAUCU (((.((((((((..........))).))))))))........--...........((((--(((......)))..............(((((........))))).....))))..... ( -25.90) >DroYak_CAF1 16156 117 + 1 GUGUGCAUCGCAAUCAAAGCUCUGCAGAUGCCACUACAUCCGGGGGAUUUCCGGGAACCGAGCCAAUUUGGCCCACCCUUUCUUUAAAUUAGCCCAAAA--CAAUACAAUGGCUAAUCU (((.((((((((..........))).)))))))).......(((((....))(((..(((((....)))))))).))).........(((((((.....--.........))))))).. ( -31.74) >consensus GUGUGCAUCGCAAUCAAAGCUCCGCAGAUGCCACUACAUCCG__CG___________________AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA_CGCAAUACAAUGGCUAAUCU (((.(((((((............)).))))))))................................(((((((((..............)))))))))..................... (-18.52 = -19.32 + 0.80)

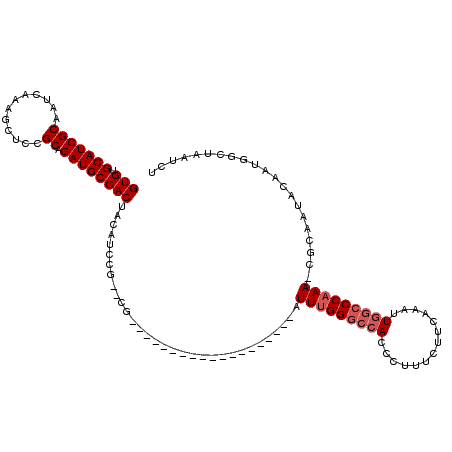
| Location | 6,324,789 – 6,324,886 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -26.23 |
| Energy contribution | -26.27 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6324789 97 - 22407834 AGAUUAGCCAUUGUAUAGCG-UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU-------------------CG--CGGAUGUAGUGGCAUCUGCGGAGCUUUGAUUGCGAUGCACAC ......((.((((((.((((-(((((((((.(((......))).))))))))))-------------------((--(((((((....)))))))))..))).....)))))))).... ( -39.50) >DroSec_CAF1 15893 97 - 1 AGAUUAGCCAUUGUAUUGCG-UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU-------------------CG--CGGAUGUAGUGGCAUCUGCGAAGCUUUGAUUGCGAUGCACAC ...........(((((((((-(((((((((.(((......))).)))))))))(-------------------((--(((((((....)))))))))).........)))))))))... ( -40.50) >DroSim_CAF1 16094 97 - 1 AGAUUAGCCAUUGUAUUGCG-UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU-------------------CG--CGGAUGUAGUGGCAUCUGCGGAGCUUUGAUUGCGAUGCACAC ...........(((((((((-(((((((((.(((......))).))))))))))-------------------((--(((((((....)))))))))...........))))))))... ( -39.90) >DroEre_CAF1 16205 115 - 1 AGAUUAGCCAUUGUAUUGCGUUUUGGGGCAAUUUGAAGAAAGGGUGGCCCGAAUCGGC--GGCUCAUAGAAAUCG--CUGAUGUAGUGGCAUCUGCAGAGCUUUGAUUGCGAUGCACAC ...........(((((((((..(..((((............((((.(((......)))--.)))).........(--(.(((((....))))).))...))))..).)))))))))... ( -37.20) >DroYak_CAF1 16156 117 - 1 AGAUUAGCCAUUGUAUUG--UUUUGGGCUAAUUUAAAGAAAGGGUGGGCCAAAUUGGCUCGGUUCCCGGAAAUCCCCCGGAUGUAGUGGCAUCUGCAGAGCUUUGAUUGCGAUGCACAC ((((..(((((((((((.--...((((..............(((((((((.....))))))...)))((....)))))))))))))))))))))(((..((.......))..))).... ( -36.40) >consensus AGAUUAGCCAUUGUAUUGCG_UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU___________________CG__CGGAUGUAGUGGCAUCUGCGGAGCUUUGAUUGCGAUGCACAC ((((..(((((((((((....(((((((((.(((......))).)))))))))..........................)))))))))))))))(((..((.......))..))).... (-26.23 = -26.27 + 0.04)

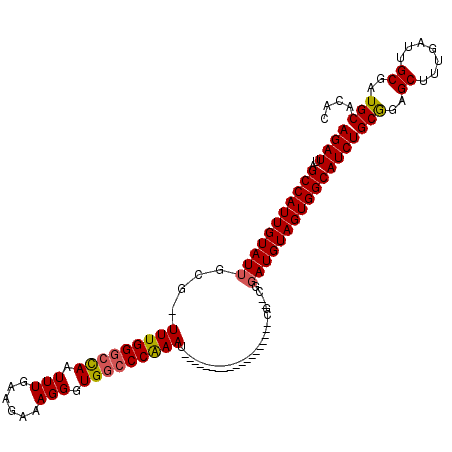
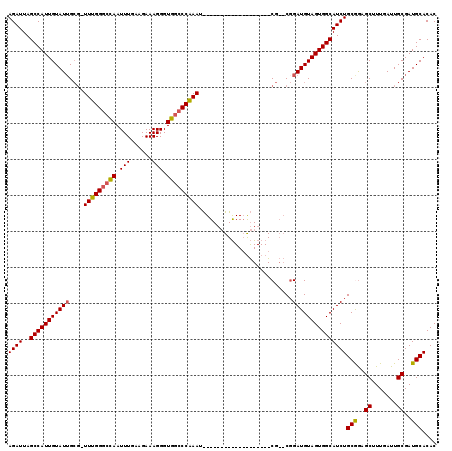
| Location | 6,324,828 – 6,324,923 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -18.00 |
| Energy contribution | -19.04 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6324828 95 + 22407834 CCG--CG-------------------AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA-CGCUAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUACAAAAGCAAACAAUG ..(--((-------------------.(((((((((..............)))))))))-))).......(((((........))))).(((((((.(....).)))))))...... ( -28.74) >DroSec_CAF1 15932 95 + 1 CCG--CG-------------------AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA-CGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUG ..(--((-------------------.(((((((((..............)))))))))-))).......(((((........)))))(((((((..((((....))))))))))). ( -31.64) >DroSim_CAF1 16133 95 + 1 CCG--CG-------------------AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA-CGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUG ..(--((-------------------.(((((((((..............)))))))))-))).......(((((........)))))(((((((..((((....))))))))))). ( -31.64) >DroEre_CAF1 16244 113 + 1 CAG--CGAUUUCUAUGAGCC--GCCGAUUCGGGCCACCCUUUCUUCAAAUUGCCCCAAAACGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUG ..(--(...(((((......--(((((...(((...))).........(((((........)))))(((((((((........)))).)))))..)))))))))).))......... ( -22.90) >DroYak_CAF1 16195 115 + 1 CCGGGGGAUUUCCGGGAACCGAGCCAAUUUGGCCCACCCUUUCUUUAAAUUAGCCCAAAA--CAAUACAAUGGCUAAUCUGUUGUCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUG ..(((((....))(((..(((((....)))))))).)))..........((((((.((((--(((((((((((.....)))))))...)))))))).)))))).............. ( -29.60) >consensus CCG__CG___________________AUUUGGGCCACCCUUUCUUCAAAUUGGCCCAAA_CGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUG ...........................(((((((((..............)))))))))...........(((((........)))))(((((((..((((....))))))))))). (-18.00 = -19.04 + 1.04)

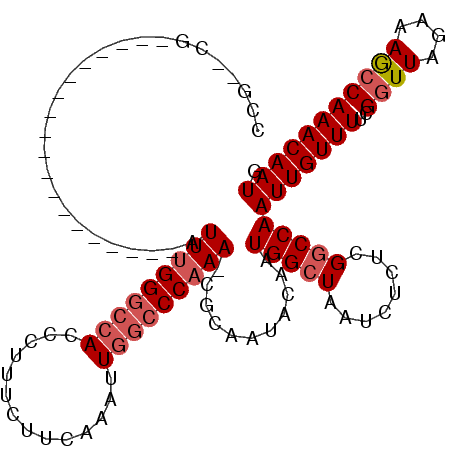
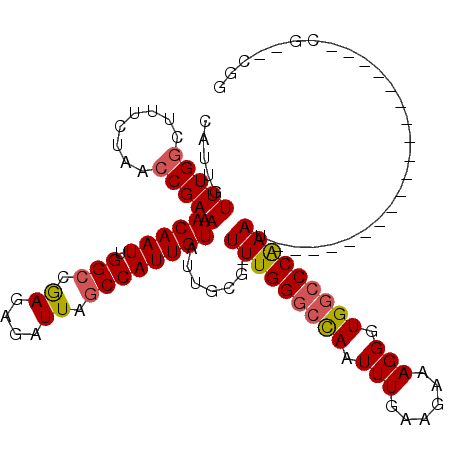
| Location | 6,324,828 – 6,324,923 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6324828 95 - 22407834 CAUUGUUUGCUUUUGUAACCGAAAACAAUUGGCCGAGAGAUUAGCCAUUGUAUAGCG-UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU-------------------CG--CGG ..(((.((((....)))).)))..(((((.(((..(....)..))))))))...(((-(((((((((.(((......))).))))))))).-------------------))--).. ( -32.00) >DroSec_CAF1 15932 95 - 1 CAUUGUUUGGCUUUCUAACCGAAAACAAUUGGCCGAGAGAUUAGCCAUUGUAUUGCG-UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU-------------------CG--CGG .......(((((((((..((((......))))...))))...)))))......((((-(((((((((.(((......))).))))))))).-------------------))--)). ( -31.90) >DroSim_CAF1 16133 95 - 1 CAUUGUUUGGCUUUCUAACCGAAAACAAUUGGCCGAGAGAUUAGCCAUUGUAUUGCG-UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU-------------------CG--CGG .......(((((((((..((((......))))...))))...)))))......((((-(((((((((.(((......))).))))))))).-------------------))--)). ( -31.90) >DroEre_CAF1 16244 113 - 1 CAUUGUUUGGCUUUCUAACCGAAAACAAUUGGCCGAGAGAUUAGCCAUUGUAUUGCGUUUUGGGGCAAUUUGAAGAAAGGGUGGCCCGAAUCGGC--GGCUCAUAGAAAUCG--CUG ........(((((((((.((.(((((...((((..(....)..))))..(.....)))))).))..............((((.(((......)))--.)))).))))))..)--)). ( -29.60) >DroYak_CAF1 16195 115 - 1 CAUUGUUUGGCUUUCUAACCGAAAACAAUUGGACAACAGAUUAGCCAUUGUAUUG--UUUUGGGCUAAUUUAAAGAAAGGGUGGGCCAAAUUGGCUCGGUUCCCGGAAAUCCCCCGG ..........((((((..((((......)))).....(((((((((...(.....--.)...)))))))))..))))))..((((((.....))))))....((((.......)))) ( -30.90) >consensus CAUUGUUUGGCUUUCUAACCGAAAACAAUUGGCCGAGAGAUUAGCCAUUGUAUUGCG_UUUGGGCCAAUUUGAAGAAAGGGUGGCCCAAAU___________________CG__CGG .....(((((........))))).(((((.(((.((....)).)))))))).......(((((((((.(((......))).)))))))))........................... (-19.62 = -19.94 + 0.32)

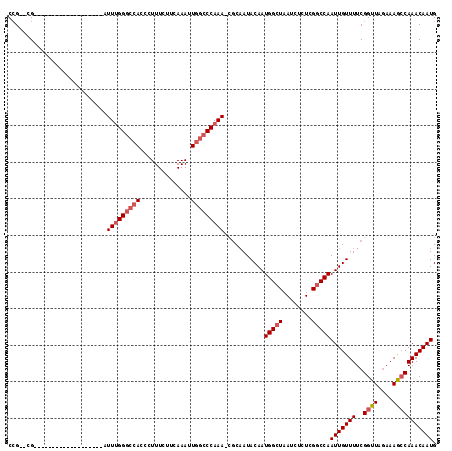
| Location | 6,324,847 – 6,324,957 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6324847 110 + 22407834 UUCUUCAAAUUGGCCCAAA-CGCUAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUACAAAAGCAAACAAUGAACAAAUGGCAAUUUUAAAAGUUGCCACUAUUGU ....(((((((((((....-.(((((...))))).........))))))))(((((.(....).)))))......)))((((.((((((((.....))))))))...)))) ( -24.74) >DroSec_CAF1 15951 109 + 1 UUCUUCAAAUUGGCCCAAA-CGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUGUACAAAUGGCAAUUAA-AAAGUUGCCACUGUUGU .........(((((.....-......(((((((((........)))).)))))((((.....))))))))).......((((.((((((((..-..))))))))...)))) ( -26.50) >DroSim_CAF1 16152 110 + 1 UUCUUCAAAUUGGCCCAAA-CGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUGUACAAAUGGCAAUUAAAAAAGUUGCCACUAUUGU .........(((((.....-......(((((((((........)))).)))))((((.....)))))))))((((((......((((((((.....)))))))).)))))) ( -25.60) >DroEre_CAF1 16280 111 + 1 UUCUUCAAAUUGCCCCAAAACGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUGUACAAACGGCAAUUAAAAUGGUCGCUUCUGUUGU .......(((((((................(((((........)))))(((((((..((((....)))))))))))........)))))))..(((((......))))).. ( -22.20) >DroYak_CAF1 16235 109 + 1 UUCUUUAAAUUAGCCCAAAA--CAAUACAAUGGCUAAUCUGUUGUCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUGUACAAAUGGCAGUUAAAAACGUCGCUGCUAUUGU .........((((((.((((--(((((((((((.....)))))))...)))))))).))))))..................(((((((((..........))))))))).. ( -25.20) >consensus UUCUUCAAAUUGGCCCAAA_CGCAAUACAAUGGCUAAUCUCUCGGCCAAUUGUUUUCGGUUAGAAAGCCAAACAAUGUACAAAUGGCAAUUAAAAAAGUUGCCACUAUUGU .....................((((((...(((((........)))))(((((((..((((....))))))))))).......((((((((.....)))))))).)))))) (-17.16 = -17.72 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:25 2006