| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,310,614 – 6,310,706 |
| Length | 92 |
| Max. P | 0.863511 |

| Location | 6,310,614 – 6,310,706 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
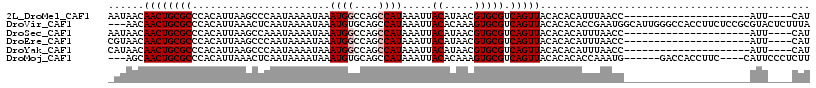
>2L_DroMel_CAF1 6310614 92 + 22407834 AUG----AAU---------------------GGUUAAAUGUGUGUAACUGACGCACGUUAUGUAAUUUAUGGCUGGCCAUUUAUUUUAUUGGGCUUAAUGUGGGCGCAGUUGUUAUU (((----(((---------------------(((((...((((((.....))))))((((((.....)))))))))))))))))...((((.((((.....)))).))))....... ( -26.90) >DroVir_CAF1 1174 114 + 1 UAAAGAGUACGCGGAGAAGGUGGCCCAAUGCCAUUCGGUGUGUGUAACUGACGCACUUUGUGUAAUUUAUGGCUGCACAUUUAUUUUAUUGAGUUUAAUGUGGGCGCAGUUGUU--- ...(((.(((((((((...(((((.....)))))(((((.......)))))....))))))))).)))(..((((((((((.((((....))))..)))))....)))))..).--- ( -30.00) >DroSec_CAF1 1847 92 + 1 AUG----AAU---------------------GGUUAAAUGUGUGUAACUGACGCACGUUAUGUAAUUUAUGGCUGGCCAUUUAUUUUAUUUGGCUUAAUGUGGGCGCAGUUGUUAUU (((----(((---------------------(((((...((((((.....))))))((((((.....))))))))))))))))).....(((((((.....)))).)))........ ( -23.80) >DroEre_CAF1 1782 92 + 1 AUG----AAU---------------------GGUUAAAUGUGUGUAACUGACGCACGUUAUGUAAUUUAUGGCUGGCCAUUUAUUUUAUUGGGCUUAAUGUGGGCGCAGUUGUUACG (((----(((---------------------(((((...((((((.....))))))((((((.....)))))))))))))))))...((((.((((.....)))).))))....... ( -26.90) >DroYak_CAF1 1777 92 + 1 AUG----AAU---------------------GGUUAAAUGUGUGUAACUGACGCACGUUAUGUAAUUUAUGGCUGGCCAUUUAUUUUAUUGGGCUUAAUGUGGGCGCAGUUGUUAUG (((----(((---------------------(((((...((((((.....))))))((((((.....)))))))))))))))))...((((.((((.....)))).))))....... ( -26.90) >DroMoj_CAF1 5867 104 + 1 AAGAGGGAAUG----GAAGGUGGUC------CAUUUGGUGUGUGUAACUGACGCACUUUGUGUAAUUUAUGGCUGCACAUUUAUUUUAUUGAGUUUAAUGUGGGCGCAGUUGCU--- ......(((((----((......))------))))).......(((((((((((.....))))........(((.((((((.((((....))))..))))))))).))))))).--- ( -27.30) >consensus AUG____AAU_____________________GGUUAAAUGUGUGUAACUGACGCACGUUAUGUAAUUUAUGGCUGGCCAUUUAUUUUAUUGGGCUUAAUGUGGGCGCAGUUGUUAU_ ....................................(((((((((.....)))))))))....(((..((((....))))..)))..((((.((((.....)))).))))....... (-14.77 = -14.93 + 0.17)

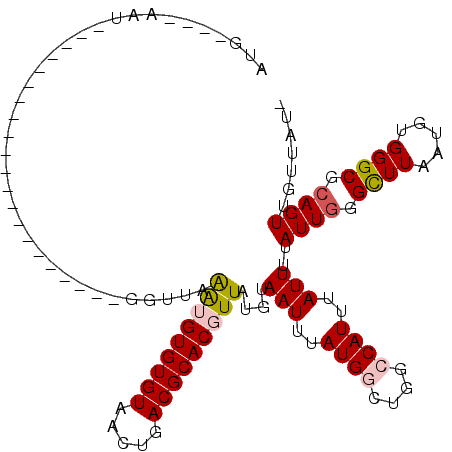
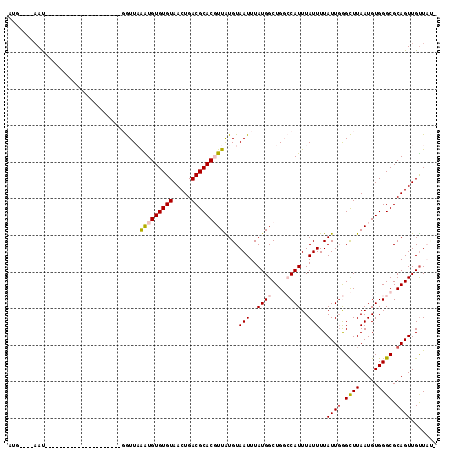
| Location | 6,310,614 – 6,310,706 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -13.43 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6310614 92 - 22407834 AAUAACAACUGCGCCCACAUUAAGCCCAAUAAAAUAAAUGGCCAGCCAUAAAUUACAUAACGUGCGUCAGUUACACACAUUUAACC---------------------AUU----CAU ......((((((((.......................((((....)))).....((.....))))).)))))..............---------------------...----... ( -11.70) >DroVir_CAF1 1174 114 - 1 ---AACAACUGCGCCCACAUUAAACUCAAUAAAAUAAAUGUGCAGCCAUAAAUUACACAAAGUGCGUCAGUUACACACACCGAAUGGCAUUGGGCCACCUUCUCCGCGUACUCUUUA ---.....((((((.........................))))))...............(((((((..((.....))...(((((((.....))))..)))...)))))))..... ( -17.81) >DroSec_CAF1 1847 92 - 1 AAUAACAACUGCGCCCACAUUAAGCCAAAUAAAAUAAAUGGCCAGCCAUAAAUUACAUAACGUGCGUCAGUUACACACAUUUAACC---------------------AUU----CAU ......((((((((.......................((((....)))).....((.....))))).)))))..............---------------------...----... ( -11.70) >DroEre_CAF1 1782 92 - 1 CGUAACAACUGCGCCCACAUUAAGCCCAAUAAAAUAAAUGGCCAGCCAUAAAUUACAUAACGUGCGUCAGUUACACACAUUUAACC---------------------AUU----CAU .(((((...(((((....(((......))).......((((....))))............)))))...)))))............---------------------...----... ( -12.30) >DroYak_CAF1 1777 92 - 1 CAUAACAACUGCGCCCACAUUAAGCCCAAUAAAAUAAAUGGCCAGCCAUAAAUUACAUAACGUGCGUCAGUUACACACAUUUAACC---------------------AUU----CAU ......((((((((.......................((((....)))).....((.....))))).)))))..............---------------------...----... ( -11.70) >DroMoj_CAF1 5867 104 - 1 ---AGCAACUGCGCCCACAUUAAACUCAAUAAAAUAAAUGUGCAGCCAUAAAUUACACAAAGUGCGUCAGUUACACACACCAAAUG------GACCACCUUC----CAUUCCCUCUU ---.(.((((((((.((((((...............))))))............((.....))))).))))).)........((((------((......))----))))....... ( -15.36) >consensus _AUAACAACUGCGCCCACAUUAAGCCCAAUAAAAUAAAUGGCCAGCCAUAAAUUACAUAACGUGCGUCAGUUACACACAUUUAACC_____________________AUU____CAU ......((((((((.......................((((....)))).....((.....))))).)))))............................................. ( -9.47 = -9.80 + 0.33)
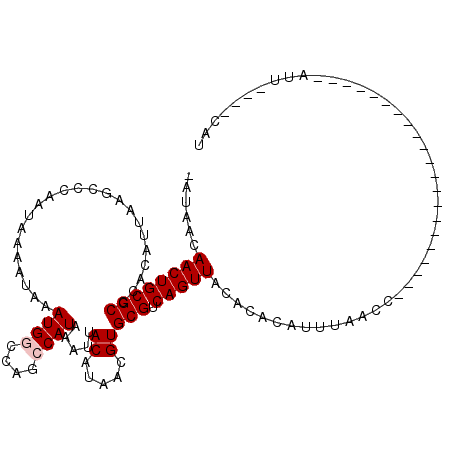
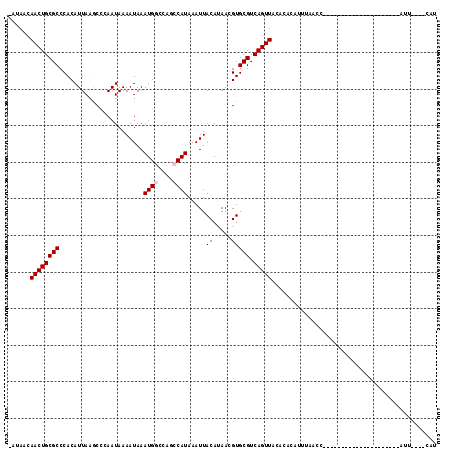
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:18 2006