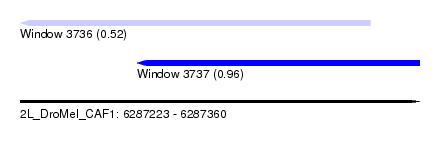
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,287,223 – 6,287,360 |
| Length | 137 |
| Max. P | 0.957606 |

| Location | 6,287,223 – 6,287,343 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
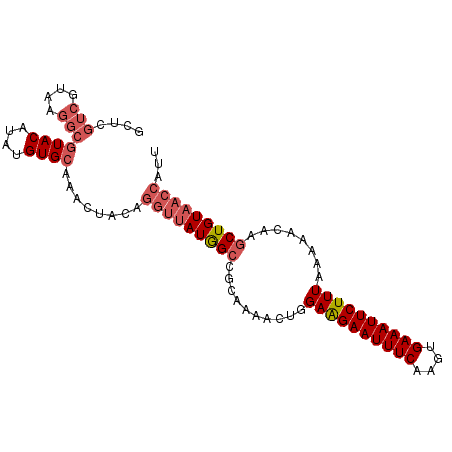
>2L_DroMel_CAF1 6287223 120 - 22407834 CAUAUGUGCAAACUACAGGUUAUGGCCGCAAAACGGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACUAGCUGUAACCAUUUUUGAAUACGGCAUGCGUUUUGGUUCAUCUCAUGCUGACU .......(((.......(((....))).(((((((.((((((((((....))))))))))........((((((..((....))..))))))...)))))))..........)))..... ( -31.00) >DroSec_CAF1 123495 120 - 1 CAUAUGUGCAAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUUUUCGAACACGGCAUGCGUUUUGGGUCAUCUUUUUCCGACU ....((((.....))))((..((((((.((((((..((((((((((....))))))))))........(((((..............)))))....))))))))))))......)).... ( -29.44) >DroSim_CAF1 138205 120 - 1 CAUAUGUGCCAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUACCCAUUUUCGAACACGGCAUGCGUUUUGGGUCAUCUUAUUCCUACC ................(((..((((((.((((((..((((((((((....))))))))))........(((((..............)))))....))))))))))))......)))... ( -29.64) >DroYak_CAF1 136438 97 - 1 CAUAUGUGCAAACUACAGGUUUUAGCCGCAAAACAGGAGGAAUUUCAAGUGAAAUUCUUUAAUAACUAGAUAUAACAAAUUU-----------------------UAUCUUAUUUCCACU .....(((.(((.....(((....))).........((((((((((....)))))))))).......(((((..........-----------------------)))))..))).))). ( -14.90) >consensus CAUAUGUGCAAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUUUUCGAACACGGCAUGCGUUUUGGGUCAUCUUAUUCCGACU ....((((.....))))(((((((((..........((((((((((....))))))))))........)))))))))........................................... (-17.71 = -18.14 + 0.44)


| Location | 6,287,263 – 6,287,360 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -19.92 |
| Energy contribution | -21.79 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6287263 97 - 22407834 GCUCGUCGUAAGGCGUACAUAUGUGCAAACUACAGGUUAUGGCCGCAAAACGGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACUAGCUGUAACCAUU ....(((....)))((((....))))........(((((((((((.....)).((((((((((....))))))))))........)))))))))... ( -30.30) >DroSec_CAF1 123535 97 - 1 GCUCGUCGUAAGGCGUACAUAUGUGCAAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUU ....(((....)))((((....))))........(((((((((..........((((((((((....))))))))))........)))))))))... ( -30.07) >DroSim_CAF1 138245 97 - 1 GCUCGUCGUAAGGCGUACAUAUGUGCCAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUACCCAUU ....(((....)))(((((..(((((((((.....))..))).))))......((((((((((....))))))))))..........)))))..... ( -25.00) >DroEre_CAF1 123171 87 - 1 GCUCCUC--------UACAUGUGUGCAAACUACAGGUUAUAGCCGCAAAACAGGAGGAAUUUCAAGUGAAAUUCUUUAAAAACUGCCUGUCAG--GU .(((((.--------....((((.((.(((.....)))...))))))....)))))(((((((....)))))))..........(((.....)--)) ( -19.50) >consensus GCUCGUCGUAAGGCGUACAUAUGUGCAAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUU ....(((....)))((((....))))........(((((((((..........((((((((((....))))))))))........)))))))))... (-19.92 = -21.79 + 1.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:07 2006