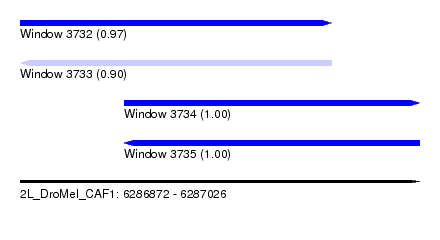
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,286,872 – 6,287,026 |
| Length | 154 |
| Max. P | 0.999961 |

| Location | 6,286,872 – 6,286,992 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -31.14 |
| Energy contribution | -30.86 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
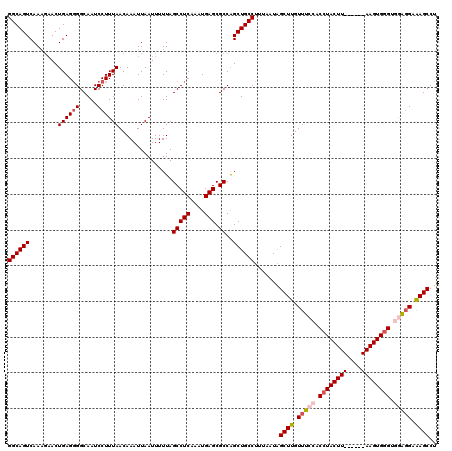
>2L_DroMel_CAF1 6286872 120 + 22407834 GCUCAGUUGGUAAAAUAUUUUGGUAAGCUCGAAGUGUUGAGGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUU (((((((((((....(((....))).(((((.....((((((((((......)))(((((....))))).................))))))).)))))))))))))........))).. ( -32.20) >DroSec_CAF1 123131 119 + 1 GCUCAGUUGGUAAAAUAUUUUGGUAAGCUCGAAGUGUUGGGGCAGUCAAAGAACUGAG-GGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUU (((((((((((....(((....))).(((((.....((((((((((......)))(((-((....)))))................))))))).)))))))))))))........))).. ( -30.60) >DroSim_CAF1 137850 117 + 1 GCUCAGUUGGUAAAAUAUUUUGGUAAGCUCGAAGUGUUGAGGCAGUCAA---ACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUU (((((((((((....(((....))).(((((.....(((((((......---..((((((.....))))))..(((......))).))))))).)))))))))))))........))).. ( -31.60) >DroEre_CAF1 122800 120 + 1 GCUCAGUUGGUAAAAUAUUUUGGUAAGCUCGAAGUGUUGAGGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUGGCUU ....(((((((....(((....))).(((((.....((((((((((......)))(((((....))))).................))))))).))))))))))))(((......))).. ( -33.60) >DroYak_CAF1 136071 120 + 1 GCUCAGUUGGUAAAAUAUUUUGGUAAGCUCGAAGUGUUGAGGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCGGCUGCCUUUAAUAGCUU (((..((((....((((((((((.....))))))))))(((((((((.......((((((.....))))))...............(((((....))).)).)))))))))))))))).. ( -32.10) >consensus GCUCAGUUGGUAAAAUAUUUUGGUAAGCUCGAAGUGUUGAGGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUU (((((((((((....(((....))).(((((.....((((((((((......)))(((((....))))).................))))))).)))))))))))))........))).. (-31.14 = -30.86 + -0.28)

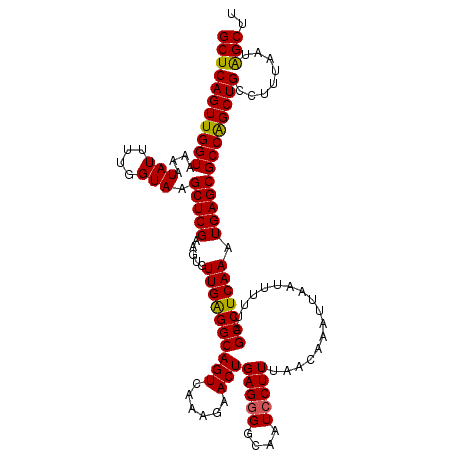
| Location | 6,286,872 – 6,286,992 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -25.44 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6286872 120 - 22407834 AAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCCUCAACACUUCGAGCUUACCAAAAUAUUUUACCAACUGAGC ..(((......)))...(((.((((..(((((((.............((((((((((((.....)))))))))))).)))))))......))))...))).................... ( -28.44) >DroSec_CAF1 123131 119 - 1 AAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCC-CUCAGUUCUUUGACUGCCCCAACACUUCGAGCUUACCAAAAUAUUUUACCAACUGAGC (((((......((((..((((.(((....)))))))...........((((((((((((..-..))))))))))))))))...........)))))........................ ( -26.63) >DroSim_CAF1 137850 117 - 1 AAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGU---UUGACUGCCUCAACACUUCGAGCUUACCAAAAUAUUUUACCAACUGAGC (((((.....((((((.((((.(((....)))))))....((((.(((....(((......)))))).---))))))))))..........)))))........................ ( -21.26) >DroEre_CAF1 122800 120 - 1 AAGCCAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCCUCAACACUUCGAGCUUACCAAAAUAUUUUACCAACUGAGC ..(((......)))...(((.((((..(((((((.............((((((((((((.....)))))))))))).)))))))......))))...))).................... ( -30.64) >DroYak_CAF1 136071 120 - 1 AAGCUAUUAAAGGCAGCCGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCCUCAACACUUCGAGCUUACCAAAAUAUUUUACCAACUGAGC ..(((......)))....((.((((..(((((((.............((((((((((((.....)))))))))))).)))))))......))))...))..................... ( -27.94) >consensus AAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCCUCAACACUUCGAGCUUACCAAAAUAUUUUACCAACUGAGC ..(((......)))....((.((((..(((((((.............((((((((((((.....)))))))))))).)))))))......))))...))..................... (-25.44 = -26.08 + 0.64)


| Location | 6,286,912 – 6,287,026 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -32.44 |
| Energy contribution | -33.64 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6286912 114 + 22407834 GGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUUGUUUCCACCUACUU------AAGUGGGCGGAGGAAAGCCU ((((((........((((((.....))))))...............(((((....))).))..)))))).......((((.(((((.(((((..------..))))).))))).)))).. ( -36.40) >DroSec_CAF1 123171 113 + 1 GGCAGUCAAAGAACUGAG-GGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUUGUUUCCACCUACUU------AAGUGGGUGGAGGAGAGCCU ((((((........((((-((....))))))...............(((((....))).))..)))))).......((((.(((((((((((..------..))))))))))).)))).. ( -40.00) >DroSim_CAF1 137890 111 + 1 GGCAGUCAA---ACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUUGUUUCCACCUACUU------AAGUGGGUGGAGGAGAGCCU ((((((...---..((((((.....))))))...............(((((....))).))..)))))).......((((.(((((((((((..------..))))))))))).)))).. ( -40.70) >DroEre_CAF1 122840 120 + 1 GGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUGGCUUGUUUCCACCUACUUACAAGUAAGUGGGAAAGGGGAAGCCU ((((((........((((((.....))))))...............(((((....))).))..))))))......((((..(..((.((((((((....))))))))...))..))))). ( -37.90) >DroYak_CAF1 136111 114 + 1 GGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCGGCUGCCUUUAAUAGCUUGUUUCCACCUACUU------AAGUGCGGAAAUGGAAGCCU (((((((.......((((((.....))))))...............(((((....))).)).)))))))...........((((((((((((..------..))).))...))))))).. ( -32.20) >consensus GGCAGUCAAAGAACUGAGGGGCAAUCCUUUAACAAAUUAAUUUUUAGCCUCAAAUGAGCGCCAGCUGCCUUUAAUAGCUUGUUUCCACCUACUU______AAGUGGGUGGAGGAAAGCCU ((((((........((((((.....))))))...............(((((....))).))..)))))).......((((.(((((.((((((((....)))))))).))))).)))).. (-32.44 = -33.64 + 1.20)


| Location | 6,286,912 – 6,287,026 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -33.88 |
| Energy contribution | -35.36 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.91 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
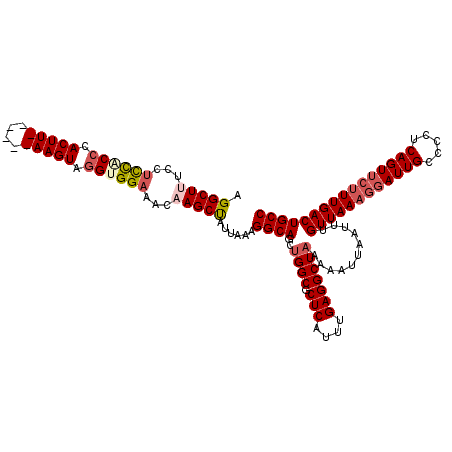
>2L_DroMel_CAF1 6286912 114 - 22407834 AGGCUUUCCUCCGCCCACUU------AAGUAGGUGGAAACAAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCC .(((((...((((((.((..------..)).))))))...)))))......((((..((((.(((....)))))))...........((((((((((((.....)))))))))))))))) ( -39.00) >DroSec_CAF1 123171 113 - 1 AGGCUCUCCUCCACCCACUU------AAGUAGGUGGAAACAAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCC-CUCAGUUCUUUGACUGCC .((((....((((((.((..------..)).))))))....))))......((((..((((.(((....)))))))...........((((((((((((..-..)))))))))))))))) ( -38.20) >DroSim_CAF1 137890 111 - 1 AGGCUCUCCUCCACCCACUU------AAGUAGGUGGAAACAAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGU---UUGACUGCC .(((..(((((((((.((..------..)).))))))((((((((......)))...((((.(((....))))))).........)))))...)))..)))...(((.---....))).. ( -33.30) >DroEre_CAF1 122840 120 - 1 AGGCUUCCCCUUUCCCACUUACUUGUAAGUAGGUGGAAACAAGCCAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCC .(((((....(((((.(((((((....)))))))))))).)))))......((((..((((.(((....)))))))...........((((((((((((.....)))))))))))))))) ( -41.20) >DroYak_CAF1 136111 114 - 1 AGGCUUCCAUUUCCGCACUU------AAGUAGGUGGAAACAAGCUAUUAAAGGCAGCCGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCC .(((((...(((((((.((.------....))))))))).)))))......((((...(((.(((....))))))............((((((((((((.....)))))))))))))))) ( -35.50) >consensus AGGCUUUCCUCCACCCACUU______AAGUAGGUGGAAACAAGCUAUUAAAGGCAGCUGGCGCUCAUUUGAGGCUAAAAAUUAAUUUGUUAAAGGAUUGCCCCUCAGUUCUUUGACUGCC .(((((...((((((.(((((....))))).))))))...)))))......((((..((((.(((....)))))))...........((((((((((((.....)))))))))))))))) (-33.88 = -35.36 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:05 2006