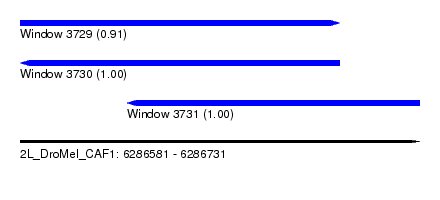
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,286,581 – 6,286,731 |
| Length | 150 |
| Max. P | 0.998395 |

| Location | 6,286,581 – 6,286,701 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.08 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
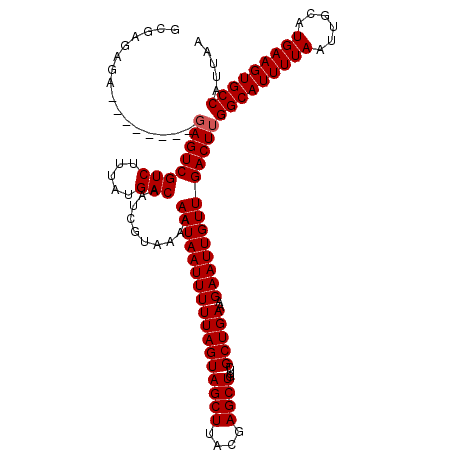
>2L_DroMel_CAF1 6286581 120 + 22407834 UUAUUUUUCAUUUAAUUUCCAAUUCCUUCGUUCGGCGCUUUUAAUGGCACUUCAUGCAAUUAAAAUGCCAAGUCAACAAUUCUUUCAGCGAACAGCUCGUAAGCUACUAAAAAUUAUUUU ............((((((...........((((((((.(((((((.(((.....))).)))))))))))..(....)............))))((((....)))).....)))))).... ( -19.10) >DroSec_CAF1 122838 120 + 1 UUAUUUUUCAUUUAAUUUCCAAUUCCUUCGUUCGGCGCUUUUAAUGGCACUUCAUGCAAUUAAAAUGCCAAGUCAACAAUUCUUUCAGCGAACAGCUCGUAAGCUACUAAAAAUUAUUUU ............((((((...........((((((((.(((((((.(((.....))).)))))))))))..(....)............))))((((....)))).....)))))).... ( -19.10) >DroSim_CAF1 137557 120 + 1 UUAUUUUUCAUUUAAUUUCCAAUUCCUUCGUUCGGCGCUUUUAAUGGCACUUCAUGCAAUUAAAAUGCCAAGUCAACAAUUCUUUCAGCGAACAGCUCGUAAGCUACUAAAAAUUAUUUU ............((((((...........((((((((.(((((((.(((.....))).)))))))))))..(....)............))))((((....)))).....)))))).... ( -19.10) >DroEre_CAF1 122497 120 + 1 UUACUUUUCAUUUAAUUUCCAAUUCCUUCGUUCGGCGCUUUUAAUGGCACUUCAUGCAAUUAAAAUGCCAAGUCAACAAUUCUUUCAGCGAGCAGCUCGUAAGCUACUAAAAAUUAUUUU ............((((((...........((((((((.(((((((.(((.....))).)))))))))))..(....)............))))((((....)))).....)))))).... ( -19.10) >DroYak_CAF1 135775 120 + 1 UUAUUUUUCAUUUAAUUUCCAAUUCCUUCGUUUGGCGCUUUUAAUGGCACUUCAUGCAAUUAAAAUGCCAAGUCAACAAUUCUUUCAGCGCACAGCUCGUAAGCUACUAAAAAUUAUUUU ............((((((...........((((((((.(((((((.(((.....))).)))))))))))))).)............(((..((.....))..))).....)))))).... ( -18.70) >consensus UUAUUUUUCAUUUAAUUUCCAAUUCCUUCGUUCGGCGCUUUUAAUGGCACUUCAUGCAAUUAAAAUGCCAAGUCAACAAUUCUUUCAGCGAACAGCUCGUAAGCUACUAAAAAUUAUUUU ............((((((...........((((((((.(((((((.(((.....))).)))))))))))..(....)............))))((((....)))).....)))))).... (-18.20 = -18.08 + -0.12)

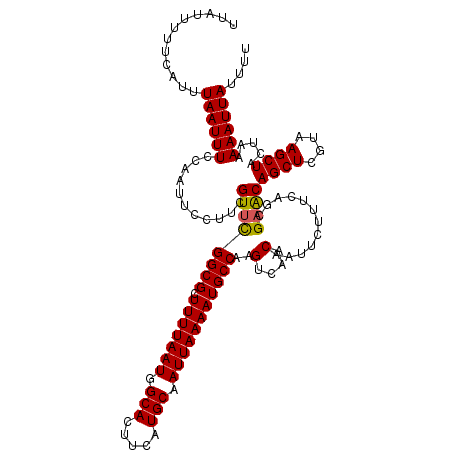

| Location | 6,286,581 – 6,286,701 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6286581 120 - 22407834 AAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAAAAGCGCCGAACGAAGGAAUUGGAAAUUAAAUGAAAAAUAA ....(((((((((((..(((.(((((.((((.......)))).)))...(((((.(((((((.((((...)))).)))))))..))))).)))))..)))))))))))............ ( -27.30) >DroSec_CAF1 122838 120 - 1 AAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAAAAGCGCCGAACGAAGGAAUUGGAAAUUAAAUGAAAAAUAA ....(((((((((((..(((.(((((.((((.......)))).)))...(((((.(((((((.((((...)))).)))))))..))))).)))))..)))))))))))............ ( -27.30) >DroSim_CAF1 137557 120 - 1 AAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAAAAGCGCCGAACGAAGGAAUUGGAAAUUAAAUGAAAAAUAA ....(((((((((((..(((.(((((.((((.......)))).)))...(((((.(((((((.((((...)))).)))))))..))))).)))))..)))))))))))............ ( -27.30) >DroEre_CAF1 122497 120 - 1 AAAAUAAUUUUUAGUAGCUUACGAGCUGCUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAAAAGCGCCGAACGAAGGAAUUGGAAAUUAAAUGAAAAGUAA ..(((((((((((((((((....)))))))......)))))))))).(((((((((((((......)))))))))..........((((.(....)..))))............)))).. ( -27.10) >DroYak_CAF1 135775 120 - 1 AAAAUAAUUUUUAGUAGCUUACGAGCUGUGCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAAAAGCGCCAAACGAAGGAAUUGGAAAUUAAAUGAAAAAUAA ....(((((((((((..(((.((...((.(((((..........((((..((((((((((......)))))))))))))).)))))))..)))))..)))))))))))............ ( -28.20) >consensus AAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAAAAGCGCCGAACGAAGGAAUUGGAAAUUAAAUGAAAAAUAA ....(((((((((((..(((.(((((.((((.......)))).)))...(((((.(((((((.((((...)))).)))))))..))))).)))))..)))))))))))............ (-26.00 = -26.24 + 0.24)

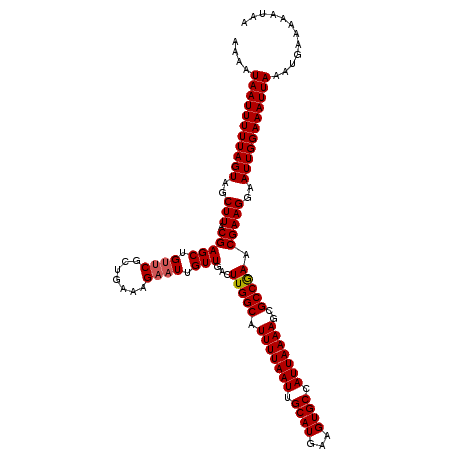
| Location | 6,286,621 – 6,286,731 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6286621 110 - 22407834 GCG--AGA--------GAGUCGUCUUUAUGACAAUCGUAAAAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAA ...--...--------((((((((.....)))..........((((((((((..((((..((.....))..)))).)))))))))))))))(((((((((......)))))))))..... ( -28.60) >DroSec_CAF1 122878 112 - 1 GCGCGAGA--------GAGUCGUCUUUAUGACAAUCGUAAAAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAA .(....).--------((((((((.....)))..........((((((((((..((((..((.....))..)))).)))))))))))))))(((((((((......)))))))))..... ( -29.00) >DroSim_CAF1 137597 112 - 1 GCGAGAGA--------GAGUCGUCUUUAUGACAAUCGUAAAAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAA ........--------((((((((.....)))..........((((((((((..((((..((.....))..)))).)))))))))))))))(((((((((......)))))))))..... ( -28.60) >DroEre_CAF1 122537 110 - 1 GCGAGAG----------AGUCGUCUUUAUGACAAUCGUAAAAAAUAAUUUUUAGUAGCUUACGAGCUGCUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAA ......(----------(((((((.....)))..........(((((((((((((((((....)))))))......)))))))))))))))(((((((((......)))))))))..... ( -31.40) >DroYak_CAF1 135815 120 - 1 GCGAGAGAGGGGGGCAGAGUCGUCUUUAUGACAAUCGUAAAAAAUAAUUUUUAGUAGCUUACGAGCUGUGCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAA ((...........)).((((((((.....)))..........((((((((((..((((.(((.....))).)))).)))))))))))))))(((((((((......)))))))))..... ( -29.80) >consensus GCGAGAGA________GAGUCGUCUUUAUGACAAUCGUAAAAAAUAAUUUUUAGUAGCUUACGAGCUGUUCGCUGAAAGAAUUGUUGACUUGGCAUUUUAAUUGCAUGAAGUGCCAUUAA ................((((((((.....)))..........(((((((((((((((((....))))....)))))..)))))))))))))(((((((((......)))))))))..... (-27.36 = -27.56 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:01 2006