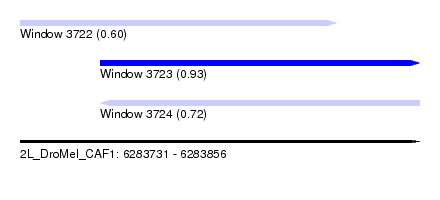
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,283,731 – 6,283,856 |
| Length | 125 |
| Max. P | 0.929714 |

| Location | 6,283,731 – 6,283,830 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
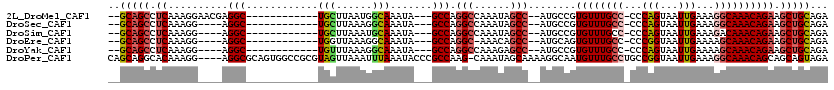
>2L_DroMel_CAF1 6283731 99 + 22407834 AAGA------CUGGGAGUCGUGCUUACGCAGUCUGCAGCUUCUGUUUGCCUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUAUUUGGCCUGGC---UAUUUGCCAUU .(((------(((.(((.....)))...))))))...(((..(((((((((((........)))-)))))))).)))..--((((....))))((((---.....)))).. ( -35.80) >DroPse_CAF1 214054 100 + 1 CAGGUCUUCUCUGGG----GUCUUCUCGCCGUC------UGCUGUUUGCCUUUCAAUUACCGGCAGGCAAACAUUGCCUUUUGCUAUUUG-CUUGGCGGGUAUUUAAAUUU .((..((......))----..)).(((((((..------.(((((((((((.((.......)).)))))))))..((.....)).....)-).)))))))........... ( -28.60) >DroSec_CAF1 120029 99 + 1 AAGA------CUGGGAGUCGUGCUUACGCAGUCUGCAGCUUCUGUUUGCCUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUAUUUGGCCUGGC---UAUUUGCCUUU .(((------(((.(((.....)))...))))))...(((..(((((((((((........)))-)))))))).)))..--((((....)))).(((---.....)))... ( -34.70) >DroSim_CAF1 134722 99 + 1 AAGA------CUGGGAGUCGUGCUUACGCAGUCUGCAGCUUCUGUUUGUCUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUAUUUGGCCUGGC---UAUUUGCAUUU .(((------(((.(((.....)))...))))))((((((..((((((.((..((.....))..-)))))))).)))((--(((((.......))))---))).))).... ( -31.60) >DroEre_CAF1 119713 98 + 1 AAGA------CUGGGAGUCGUGCUUACGCAGUCUGCAGCUUCUGUUUGCUUUUCAAUUACCGGG-GGCAAACACUGCAU--GGCUGUUU-GCCUGGC---UAUUUGCCUUU .(((------(((.(((.....)))...))))))((((....(((((((((((........)))-))))))))))))..--(((.....-))).(((---.....)))... ( -32.60) >DroPer_CAF1 213341 106 + 1 CAGGUCUUCUCUGGG----GUCUUCUCGCCGUCUACUGCUGCUGUUUGCCUUUCAAUUACCGGCAGGCAAACAUUGCCUUUUGCUAUUUG-CUUGGCGGGUAUUUAAAUUU .((..((......))----..)).(((((((......((...(((((((((.((.......)).)))))))))..)).....((.....)-).)))))))........... ( -29.20) >consensus AAGA______CUGGGAGUCGUGCUUACGCAGUCUGCAGCUUCUGUUUGCCUUUCAAUUACCGGG_GGCAAACACGGCAU__GGCUAUUUGGCCUGGC___UAUUUGCAUUU ..........(..((....(((....))).(((....((...((((((((.(((.......))).))))))))..))....))).......))..)............... (-15.86 = -16.70 + 0.84)

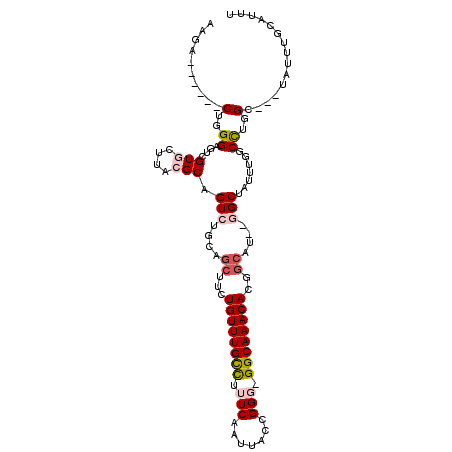
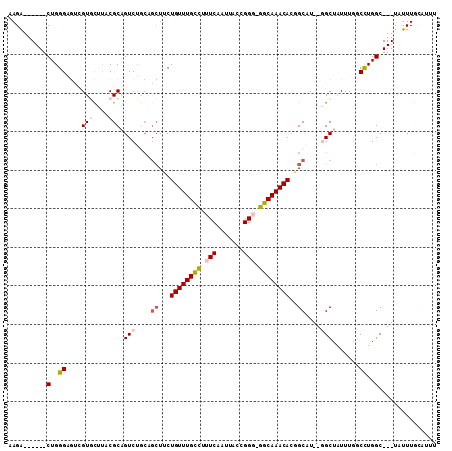
| Location | 6,283,756 – 6,283,856 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -25.57 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
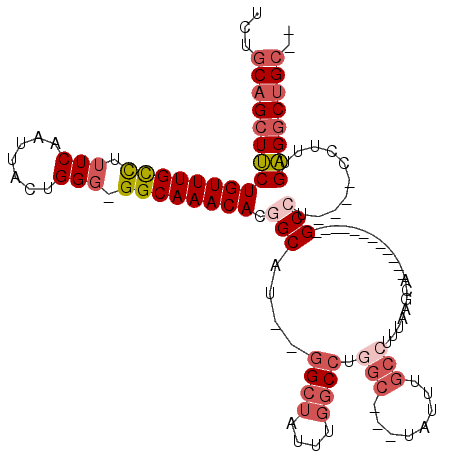
>2L_DroMel_CAF1 6283756 100 + 22407834 UCUGCAGCUUCUGUUUGCCUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUAUUUGGCCUGGC---UAUUUGCCAUUAAGCA------------GCCUCGUUCCUUUGAGGCUGC-- ......(((..(((((((((((........)))-)))))))).)))..--((((....))))((((---.....))))....(((------------((((((......)))))))))-- ( -42.80) >DroSec_CAF1 120054 96 + 1 UCUGCAGCUUCUGUUUGCCUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUAUUUGGCCUGGC---UAUUUGCCUUUAAGCA------------GCCU----CCUUUGAGGCUGC-- ...(((((((((((((((((((........)))-)))))))).(((..--((((....)))).(((---.....)))........------------))).----.....))))))))-- ( -39.90) >DroSim_CAF1 134747 96 + 1 UCUGCAGCUUCUGUUUGUCUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUAUUUGGCCUGGC---UAUUUGCAUUUAAGCA------------GCCU----CCUUUGAGGCUGC-- ..(((((((..((((((.((..((.....))..-)))))))).)))((--(((((.......))))---))).)))).....(((------------((((----(....))))))))-- ( -37.30) >DroEre_CAF1 119738 95 + 1 UCUGCAGCUUCUGUUUGCUUUUCAAUUACCGGG-GGCAAACACUGCAU--GGCUGUUU-GCCUGGC---UAUUUGCCUUUAACCA------------GCCU----CCUUUGAGGCUGC-- ...(((((((((((((((((((........)))-))))))))......--(((((.((-(...(((---.....)))..))).))------------))).----.....))))))))-- ( -36.20) >DroYak_CAF1 133135 96 + 1 UCUGCAGCUUCUGUUUGCUUUUCAAUUACUGGG-GGCAAACACGGCAU--GGCUCUUUGGCCUGGC---UAUUUGCCUUUAAACA------------GCCU----CCUUUGAGGCUGC-- ...(((((((((((((((.(..((.....))..-)))))))).(((..--((((....)))).(((---.....)))........------------))).----.....))))))))-- ( -37.40) >DroPer_CAF1 213368 115 + 1 UCUACUGCUGCUGUUUGCCUUUCAAUUACCGGCAGGCAAACAUUGCCUUUUGCUAUUUG-CUUGGCGGGUAUUUAAAUUUAACUACGCGGCCACUGCGCCU----CCUUUGUGCCUGCUG ......((((((((((((((.((.......)).))))))))).(((((...((((....-..)))))))))...............)))))....((((..----.....))))...... ( -31.30) >consensus UCUGCAGCUUCUGUUUGCCUUUCAAUUACUGGG_GGCAAACACGGCAU__GGCUAUUUGGCCUGGC___UAUUUGCCUUUAAGCA____________GCCU____CCUUUGAGGCUGC__ ...((((((((((((((((.(((.......))).)))))))).(((....((((....)))).(((........)))....................)))..........)))))))).. (-25.57 = -27.10 + 1.53)
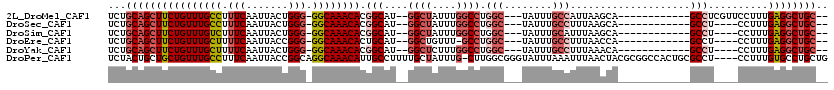
| Location | 6,283,756 – 6,283,856 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
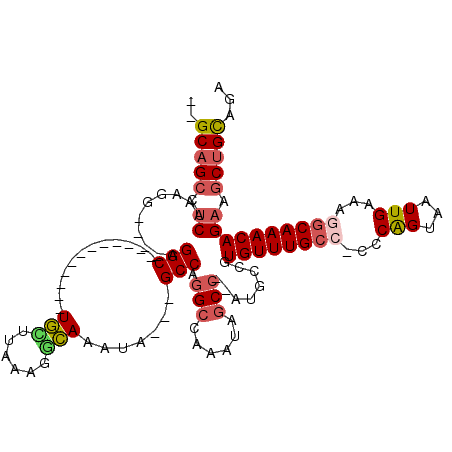
>2L_DroMel_CAF1 6283756 100 - 22407834 --GCAGCCUCAAAGGAACGAGGC------------UGCUUAAUGGCAAAUA---GCCAGGCCAAAUAGCC--AUGCCGUGUUUGCC-CCCAGUAAUUGAAAGGCAAACAGAAGCUGCAGA --((((((((........)))))------------)))....((((.....---))))(((......)))--.(((.(((((((((-..(((...)))...)))))))....)).))).. ( -36.50) >DroSec_CAF1 120054 96 - 1 --GCAGCCUCAAAGG----AGGC------------UGCUUAAAGGCAAAUA---GCCAGGCCAAAUAGCC--AUGCCGUGUUUGCC-CCCAGUAAUUGAAAGGCAAACAGAAGCUGCAGA --((((((((....)----))))------------))).....(((.....---))).(((......)))--.(((.(((((((((-..(((...)))...)))))))....)).))).. ( -35.60) >DroSim_CAF1 134747 96 - 1 --GCAGCCUCAAAGG----AGGC------------UGCUUAAAUGCAAAUA---GCCAGGCCAAAUAGCC--AUGCCGUGUUUGCC-CCCAGUAAUUGAAAGACAAACAGAAGCUGCAGA --((((((((....)----))))------------)))......(((((((---(.(((((......)))--.)).).))))))).-..((((..(((.........)))..)))).... ( -28.90) >DroEre_CAF1 119738 95 - 1 --GCAGCCUCAAAGG----AGGC------------UGGUUAAAGGCAAAUA---GCCAGGC-AAACAGCC--AUGCAGUGUUUGCC-CCCGGUAAUUGAAAAGCAAACAGAAGCUGCAGA --.(((((((....)----))))------------))......(((.....---))).(((-.....)))--.((((((((((((.-...............))))))....)))))).. ( -33.89) >DroYak_CAF1 133135 96 - 1 --GCAGCCUCAAAGG----AGGC------------UGUUUAAAGGCAAAUA---GCCAGGCCAAAGAGCC--AUGCCGUGUUUGCC-CCCAGUAAUUGAAAAGCAAACAGAAGCUGCAGA --(((((.((...((----.(((------------(((((......)))))---))).(((......)))--...)).(((((((.-...............))))))))).)))))... ( -33.29) >DroPer_CAF1 213368 115 - 1 CAGCAGGCACAAAGG----AGGCGCAGUGGCCGCGUAGUUAAAUUUAAAUACCCGCCAAG-CAAAUAGCAAAAGGCAAUGUUUGCCUGCCGGUAAUUGAAAGGCAAACAGCAGCAGUAGA ..((.(((.......----.(((......)))(.(((.(((....))).))).))))..(-(.....((.....))..(((((((((..((.....))..))))))))))).))...... ( -29.90) >consensus __GCAGCCUCAAAGG____AGGC____________UGCUUAAAGGCAAAUA___GCCAGGCCAAAUAGCC__AUGCCGUGUUUGCC_CCCAGUAAUUGAAAGGCAAACAGAAGCUGCAGA ..(((((.((..........(((............(((......))).......))).(((......)))........((((((((...(((...)))...)))))))))).)))))... (-19.27 = -19.83 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:55 2006