
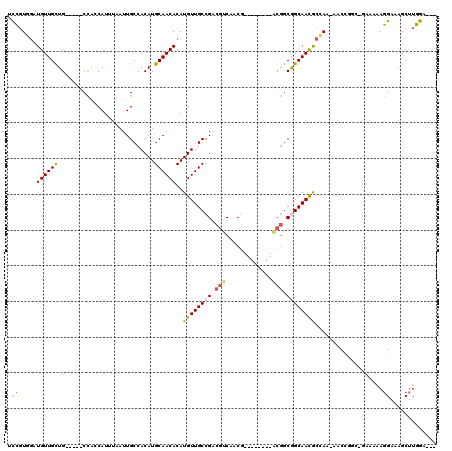
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,270,539 – 6,270,716 |
| Length | 177 |
| Max. P | 0.997412 |

| Location | 6,270,539 – 6,270,646 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -18.26 |
| Energy contribution | -17.85 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6270539 107 + 22407834 CCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCG--------ACGGCGGCAACGCCAA-AACCGGC-GAAAAAGGAAAGCUUGGA--- ((((((..((((((((((.(..............).)))).))))))))))...((....((((((--------..((((....))))..-...))))-))....)).......)).--- ( -35.64) >DroPse_CAF1 190003 103 + 1 UCCGUUGCUGUUGCUG-----CCGGCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGCCAACGCCGCUGUGGCGGCAGCAACACAAA-A--------AAAGAGGACAGCUUGGA--- (((.(((.((((((((-----((((((......))))....(((((....)))))....((((.((....))))))))))))))))))).-.--------...(((.....))))))--- ( -39.80) >DroEre_CAF1 105926 107 + 1 ACCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCG--------ACGGCGGCAAUGCCAA-AACCGGC-GAAAAAGGAAAGCUUGGA--- .(((((..((((((((((.(..............).)))).)))))))))(((((((.((((...)--------))).)))))))(((..-....)))-......))..........--- ( -31.94) >DroWil_CAF1 177265 106 + 1 UUUCUUCGUGUUGUUG-----CUGGCAUUUAAUUGCCACAUGCAACACAUGUUGCCGGCAACAACA--------GCAACGGCAACGCCAA-CGCUGGAUAAAAAAAGACAGCAUGAAAGA ((((...(((((((((-----.(((((......))))))).)))))))..(((((((((.......--------))..))))))).....-.((((..(......)..))))..)))).. ( -36.60) >DroYak_CAF1 119331 108 + 1 CCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCG--------ACGGCGGCAACGCCAAAAACCGGC-GAAAAAGGAAAGCUUGGA--- ((((((..((((((((((.(..............).)))).))))))))))...((....((((((--------..((((....))))......))))-))....)).......)).--- ( -35.04) >DroPer_CAF1 189602 100 + 1 UCCGUUGCUGUUGCUG-----CCGGCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGCCAACGCCGCUGUGGCGGCAGCAACACAAA-A--------A---AGGACAGCUUGGA--- (((.(((.((((((((-----((((((......))))....(((((....)))))....((((.((....))))))))))))))))))).-.--------.---.))).........--- ( -41.20) >consensus UCCGUGGAUGUUGCUG_____CCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCAACG________ACGGCGGCAACGCCAA_AACCGGC_GAAAAAGGAAAGCUUGGA___ .((.....((((((...........................))))))..((((((((.(((.............))).))))))))...................))............. (-18.26 = -17.85 + -0.41)


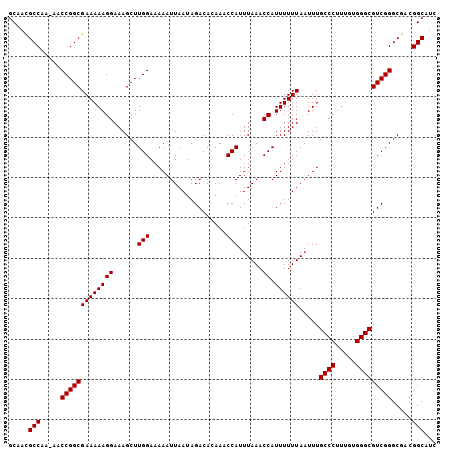
| Location | 6,270,539 – 6,270,646 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -38.91 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6270539 107 - 22407834 ---UCCAAGCUUUCCUUUUUC-GCCGGUU-UUGGCGUUGCCGCCGU--------CGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGG ---.....(((..........-((((...-.(((((((((((....--------)))))))))))(((((....)))))...)))).........(((....))))))............ ( -38.20) >DroPse_CAF1 190003 103 - 1 ---UCCAAGCUGUCCUCUUU--------U-UUUGUGUUGCUGCCGCCACAGCGGCGUUGGCGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCGG-----CAGCAACAGCAACGGA ---(((..(((((.......--------.-...(((((((((.((((((......).))))).))))))))).((((((....((((......)))).)-----))))))))))...))) ( -42.00) >DroEre_CAF1 105926 107 - 1 ---UCCAAGCUUUCCUUUUUC-GCCGGUU-UUGGCAUUGCCGCCGU--------CGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGU ---..................-((((...-...((((((((((((.--------((....)).)))).(((((.....)))))))))))......(((....))).))........)))) ( -37.74) >DroWil_CAF1 177265 106 - 1 UCUUUCAUGCUGUCUUUUUUUAUCCAGCG-UUGGCGUUGCCGUUGC--------UGUUGUUGCCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCAG-----CAACAACACGAAGAAA ............(((((........((((-.((((...)))).)))--------)((((((((..(((((....)))))...(((((......))))))-----)))))))..))))).. ( -35.30) >DroYak_CAF1 119331 108 - 1 ---UCCAAGCUUUCCUUUUUC-GCCGGUUUUUGGCGUUGCCGCCGU--------CGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGG ---.....(((..........-((((.....(((((((((((....--------)))))))))))(((((....)))))...)))).........(((....))))))............ ( -38.20) >DroPer_CAF1 189602 100 - 1 ---UCCAAGCUGUCCU---U--------U-UUUGUGUUGCUGCCGCCACAGCGGCGUUGGCGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCGG-----CAGCAACAGCAACGGA ---(((..(((((...---.--------.-...(((((((((.((((((......).))))).))))))))).((((((....((((......)))).)-----))))))))))...))) ( -42.00) >consensus ___UCCAAGCUGUCCUUUUUC_GCCGGUU_UUGGCGUUGCCGCCGC________CGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCGG_____CAGCAACAUCAACGGA ....((.............................(((((((.(((.............))).)))))))..(((((((.((......................)))))))))....)). (-18.75 = -18.55 + -0.19)

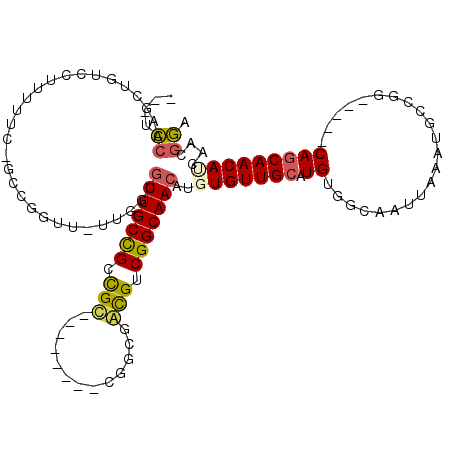

| Location | 6,270,579 – 6,270,682 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 99.23 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -25.07 |
| Energy contribution | -24.91 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6270579 103 + 22407834 UGCAACACAUGUUGCCGACGUCGCCGACGGCGGCAACGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUU .(((((....))))).....((((((..((((....))))..-...))))))....((......(((...................)))......))....... ( -26.51) >DroSec_CAF1 106593 103 + 1 UGCAACACAUGUUGCCGACGUCGCCGACGGCGGCAACGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUU .(((((....))))).....((((((..((((....))))..-...))))))....((......(((...................)))......))....... ( -26.51) >DroSim_CAF1 121355 103 + 1 UGCAACACAUGUUGCCGACGUCGCCGACGGCGGCAACGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUU .(((((....))))).....((((((..((((....))))..-...))))))....((......(((...................)))......))....... ( -26.51) >DroEre_CAF1 105966 103 + 1 UGCAACACAUGUUGCCGACGUCGCCGACGGCGGCAAUGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUU .(((((....)))))....((((((...))))))...(((..-....)))((((((((......(((...................)))......)).)))))) ( -22.41) >DroYak_CAF1 119371 104 + 1 UGCAACACAUGUUGCCGACGUCGCCGACGGCGGCAACGCCAAAAACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUU .(((((....))))).....((((((..((((....))))......))))))....((......(((...................)))......))....... ( -25.91) >consensus UGCAACACAUGUUGCCGACGUCGCCGACGGCGGCAACGCCAA_AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUU .(((((....))))).....((((((..((((....))))......))))))....((......(((...................)))......))....... (-25.07 = -24.91 + -0.16)


| Location | 6,270,579 – 6,270,682 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 99.23 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6270579 103 - 22407834 AAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCGUUGCCGCCGUCGGCGACGUCGGCAACAUGUGUUGCA .((((.((((..(((.((((((.(.............))))))).....)))..)))).))-))((((((((((....)))))))))).(((((....))))). ( -30.52) >DroSec_CAF1 106593 103 - 1 AAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCGUUGCCGCCGUCGGCGACGUCGGCAACAUGUGUUGCA .((((.((((..(((.((((((.(.............))))))).....)))..)))).))-))((((((((((....)))))))))).(((((....))))). ( -30.52) >DroSim_CAF1 121355 103 - 1 AAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCGUUGCCGCCGUCGGCGACGUCGGCAACAUGUGUUGCA .((((.((((..(((.((((((.(.............))))))).....)))..)))).))-))((((((((((....)))))))))).(((((....))))). ( -30.52) >DroEre_CAF1 105966 103 - 1 AAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCAUUGCCGCCGUCGGCGACGUCGGCAACAUGUGUUGCA .......((.......((((((.(.............)))))))..))....(((((((..-.((((......))))))))))).....(((((....))))). ( -24.82) >DroYak_CAF1 119371 104 - 1 AAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUUUUUGGCGUUGCCGCCGUCGGCGACGUCGGCAACAUGUGUUGCA (((((.((((..(((.((((((.(.............))))))).....)))..)))).)))))((((((((((....)))))))))).(((((....))))). ( -31.12) >consensus AAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU_UUGGCGUUGCCGCCGUCGGCGACGUCGGCAACAUGUGUUGCA ......((((..(((.((((((.(.............))))))).....)))..)))).....(((((((((((....)))))))))))(((((....))))). (-28.70 = -28.90 + 0.20)



| Location | 6,270,611 – 6,270,716 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 98.29 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -26.05 |
| Energy contribution | -26.05 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6270611 105 + 22407834 GCAACGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUUAAUUUGCCCUUUGUGGGCGUCGGGCGACGGCAUU .....(((..-..(((((((((((((......(((...................)))......)).)))))).....((((.....))))))))).....)))... ( -25.71) >DroSec_CAF1 106625 105 + 1 GCAACGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUUAAUUUGCCCUUUGUGGGCGUCGGGCGACGGCAUC .....(((..-..(((((((((((((......(((...................)))......)).)))))).....((((.....))))))))).....)))... ( -25.71) >DroSim_CAF1 121387 105 + 1 GCAACGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUUAAUUUGCCCUUUGUGGGCGUCGGGCGACGGCAUC .....(((..-..(((((((((((((......(((...................)))......)).)))))).....((((.....))))))))).....)))... ( -25.71) >DroEre_CAF1 105998 105 + 1 GCAAUGCCAA-AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUUAAUUUGCCCUUUGUGGGCGUCGGGCAACGGCAUC ...(((((..-..(((((((((((((......(((...................)))......)).)))))).....((((.....))))))))).....))))). ( -26.81) >DroYak_CAF1 119403 106 + 1 GCAACGCCAAAAACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUUAAUUUGCCCUUUGUGGGCGUCGGGCAACGGCAUC .....(((.....(((((((((((((......(((...................)))......)).)))))).....((((.....))))))))).....)))... ( -26.11) >consensus GCAACGCCAA_AACCGGCGAAAAAGGAAAGCUUGGAAAAAUUAAUAGACACAAACCAUUUAAACCAUUUUUUAAUUUGCCCUUUGUGGGCGUCGGGCGACGGCAUC .....(((.....(((((((((((((......(((...................)))......)).)))))).....((((.....))))))))).....)))... (-26.05 = -26.05 + 0.00)


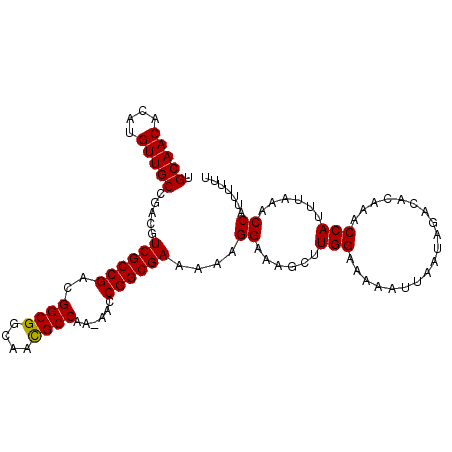
| Location | 6,270,611 – 6,270,716 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 98.29 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -26.02 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6270611 105 - 22407834 AAUGCCGUCGCCCGACGCCCACAAAGGGCAAAUUAAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCGUUGC (((((((..(((....((((.....))))............(((..(((.((((((.(.............))))))).....)))..)))))).-.))))))).. ( -26.32) >DroSec_CAF1 106625 105 - 1 GAUGCCGUCGCCCGACGCCCACAAAGGGCAAAUUAAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCGUUGC (((((((..(((....((((.....))))............(((..(((.((((((.(.............))))))).....)))..)))))).-.))))))).. ( -26.72) >DroSim_CAF1 121387 105 - 1 GAUGCCGUCGCCCGACGCCCACAAAGGGCAAAUUAAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCGUUGC (((((((..(((....((((.....))))............(((..(((.((((((.(.............))))))).....)))..)))))).-.))))))).. ( -26.72) >DroEre_CAF1 105998 105 - 1 GAUGCCGUUGCCCGACGCCCACAAAGGGCAAAUUAAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU-UUGGCAUUGC (((((((..(((....((((.....))))............(((..(((.((((((.(.............))))))).....)))..)))))).-.))))))).. ( -28.02) >DroYak_CAF1 119403 106 - 1 GAUGCCGUUGCCCGACGCCCACAAAGGGCAAAUUAAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUUUUUGGCGUUGC (((((((..(((....((((.....))))............(((..(((.((((((.(.............))))))).....)))..))))))...))))))).. ( -26.12) >consensus GAUGCCGUCGCCCGACGCCCACAAAGGGCAAAUUAAAAAAUGGUUUAAAUGGUUUGUGUCUAUUAAUUUUUCCAAGCUUUCCUUUUUCGCCGGUU_UUGGCGUUGC (((((((..(((....((((.....))))............(((..(((.((((((.(.............))))))).....)))..))))))...))))))).. (-26.02 = -25.70 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:49 2006