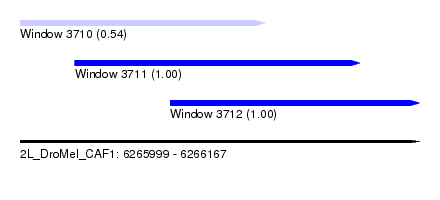
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,265,999 – 6,266,167 |
| Length | 168 |
| Max. P | 0.999963 |

| Location | 6,265,999 – 6,266,102 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6265999 103 + 22407834 ------UAAAAAGAUUUACAUCACUUUAAACUCAUCAACUCAUCAUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUG ------......(((.................(((((........)))))(((..((.....)).((((..((((....))))..))))......))))))........ ( -19.70) >DroSec_CAF1 102055 100 + 1 ------CAAAAAGAUUUACAUCACUUUAAACUCAUCAGCUC---AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUG ------......(((.................(((((....---.)))))(((..((.....)).((((..((((....))))..))))......))))))........ ( -20.50) >DroSim_CAF1 116822 106 + 1 AUUAUAUAUAAAGAUUUACAUCACUUUAAACUCAUCAGCUC---AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUG ............(((.................(((((....---.)))))(((..((.....)).((((..((((....))))..))))......))))))........ ( -20.50) >DroEre_CAF1 101333 100 + 1 ------CAACAAGACUUACAUCACUUUAAACCCAUCAGCUC---AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCAGCGUCAUUAAAUG ------......(((................((((((....---.))))))...........((.((((..((((....))))..))))....))...)))........ ( -19.60) >DroYak_CAF1 114771 100 + 1 ------GAACAAGAUUUACAUCACUUUAAACUCAUCAGCUC---AUGAUAGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCAGCGUCAUUAAAUG ------......(((..................((((....---.)))).((...((.....)).((((..((((....))))..))))....))...)))........ ( -13.40) >consensus ______CAAAAAGAUUUACAUCACUUUAAACUCAUCAGCUC___AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUG ............(((.................(((((........)))))(((..((.....)).((((..((((....))))..))))......))))))........ (-17.34 = -17.78 + 0.44)

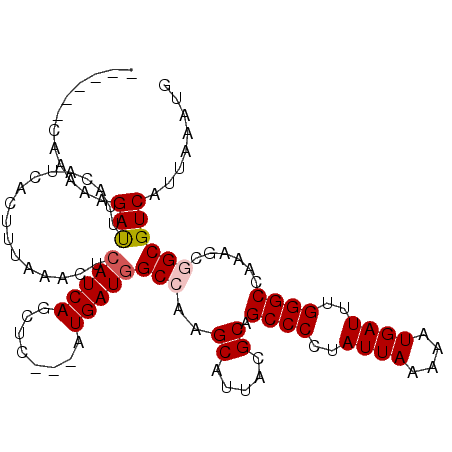
| Location | 6,266,022 – 6,266,142 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.00 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6266022 120 + 22407834 ACUCAUCAACUCAUCAUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCC ................((((((((..((.....)).((((..((((....))))..))))....((((((((........))))))))...........))))))))............. ( -34.30) >DroSec_CAF1 102078 117 + 1 ACUCAUCAGCUC---AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCC ........(((.---.((((((((..((.....)).((((..((((....))))..))))....((((((((........))))))))...........)))))))).)))......... ( -36.60) >DroSim_CAF1 116851 117 + 1 ACUCAUCAGCUC---AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCC ........(((.---.((((((((..((.....)).((((..((((....))))..))))....((((((((........))))))))...........)))))))).)))......... ( -36.60) >DroEre_CAF1 101356 116 + 1 ACCCAUCAGCUC---AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCAGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAAC-C ........(((.---.((((((((..((.....)).((((..((((....))))..))))....((((((((........))))))))...........)))))))).))).......-. ( -37.40) >DroYak_CAF1 114794 117 + 1 ACUCAUCAGCUC---AUGAUAGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCAGCGUCAUUAAAUGGACGUUGCGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCC .(((((((....---.))))......((.....)).((((..((((....))))..))))....((((((((........)))))))))))....(((((((.......))))))).... ( -34.00) >consensus ACUCAUCAGCUC___AUGAUGGCCAAGCAUUACGCAGCCCCUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCC ................((((((((..((.....)).((((..((((....))))..))))....((((((((........))))))))...........))))))))............. (-34.20 = -34.00 + -0.20)

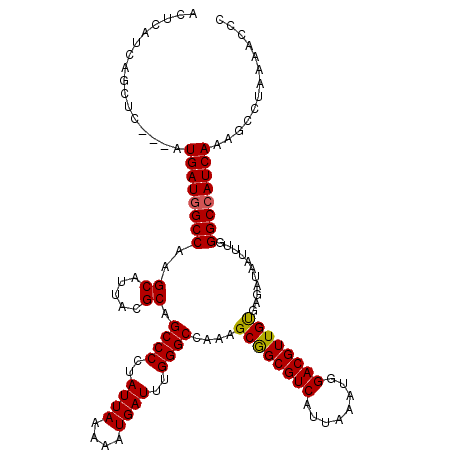
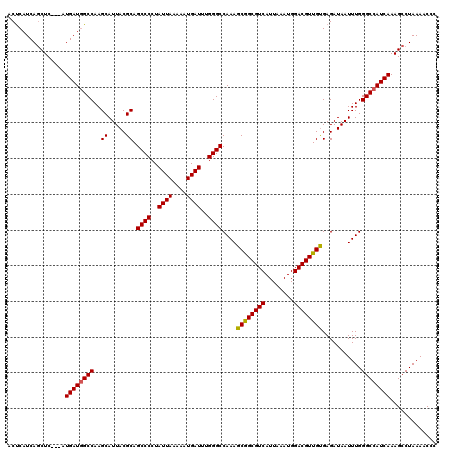
| Location | 6,266,062 – 6,266,167 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -30.40 |
| Energy contribution | -29.84 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
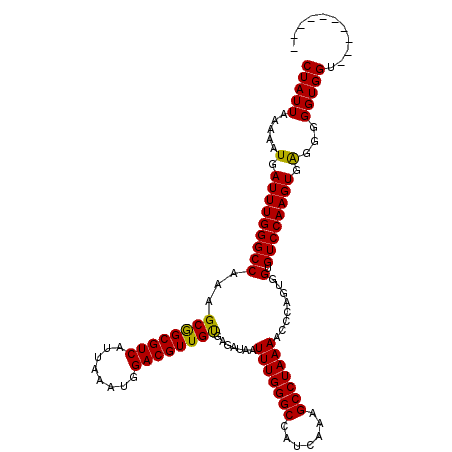
>2L_DroMel_CAF1 6266062 105 + 22407834 CUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCCAAUGGUGUCCAAGUUGGAGGGUGGU---------- (((((.....((((((((((....((((((((........)))))))).......(((((((.......)))))))(((....)))))))))))))...))))).---------- ( -30.90) >DroSec_CAF1 102115 105 + 1 CUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCCAGUGGUGUCCAAGUGAGGGGGUGGU---------- (((((.....(.((((((((....((((((((........)))))))).......(((((((.......)))))))(((....))))))))))).)...))))).---------- ( -30.30) >DroSim_CAF1 116888 105 + 1 CUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCCAGUGGUGUCCAAGUGAGGGGGUGGU---------- (((((.....(.((((((((....((((((((........)))))))).......(((((((.......)))))))(((....))))))))))).)...))))).---------- ( -30.30) >DroEre_CAF1 101393 114 + 1 CUAUUAAAAAUGAUUUGGGCCAAAGCAGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAAC-CAGUGGUGUCCAAGUAAGGGGGUGGUGAUUCUCGGC ............(((((((((...((((((((........)))))))).......(((((((.......)))))))..-.....).))))))))..((((((....))))))... ( -31.80) >DroYak_CAF1 114831 105 + 1 CUAUUAAAAAUGAUUUGGGCCAAAGCAGCGUCAUUAAAUGGACGUUGCGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCCAGUGGUGUCCAAGUGAGGGGGUGGU---------- (((((.....(.((((((((....((((((((........)))))))).......(((((((.......)))))))(((....))))))))))).)...))))).---------- ( -33.40) >consensus CUAUUAAAAAUGAUUUGGGCCAAAGCGGCGUCAUUAAAUGGACGUUGUGAGAUAAUUUGGGCCAUCAAAGCCUAAAACCCAGUGGUGUCCAAGUGAGGGGGUGGU__________ (((((.....(.(((((((((...((((((((........)))))))).......(((((((.......)))))))........).)))))))).)...)))))........... (-30.40 = -29.84 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:44 2006