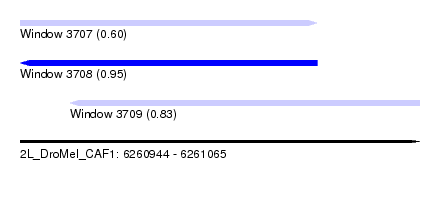
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,260,944 – 6,261,065 |
| Length | 121 |
| Max. P | 0.951668 |

| Location | 6,260,944 – 6,261,034 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
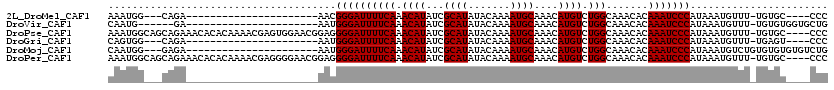
>2L_DroMel_CAF1 6260944 90 + 22407834 AAAUGG---CAGA----------------------AACGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUUU-UGUGC----CCC ....((---((((----------------------(((((((((((((.((((...((((.......))))....)))).))).......)))))))......))))-).)))----).. ( -18.71) >DroVir_CAF1 122262 91 + 1 CAAUG------GA----------------------AAUGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUUU-UGUGUGGUGCUG ....(------((----------------------.(((.(((..........)))((((.......))))...))).)))((((.(((((((............))-)))))..)))). ( -17.20) >DroPse_CAF1 172378 115 + 1 AAAUGGCAGCAGAAACACACAAAACGAGUGGAACGGAGGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUUU-UGUGC----CCC ....((((...(((((((((.......)))........((((((((((.((((...((((.......))))....)))).))).......))))))).....)))))-).)))----).. ( -24.51) >DroGri_CAF1 105118 90 + 1 CAGUGG---CAGA----------------------AAUGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUUU-UGAGU----CCC (((.((---((..----------------------.((((((((((((.((((...((((.......))))....)))).))).......)))))))))...)))))-))...----... ( -17.91) >DroMoj_CAF1 154596 95 + 1 CAAUGG---GAGA----------------------AAUGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUCUGUGUGUGUGUCUG ..((((---(...----------------------.((((((((((((.((((...((((.......))))....)))).))).......))))))))).....)))))........... ( -16.61) >DroPer_CAF1 172835 115 + 1 AAAUGGCAGCAGAAACACACAAAACGAGGGGAACGGAGGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUUU-UGUGC----CCC ....((((...((((((...........(....)....((((((((((.((((...((((.......))))....)))).))).......))))))).....)))))-).)))----).. ( -22.81) >consensus AAAUGG___CAGA______________________AAUGGGAUUUUCAAACAUAUCGCAUAUACAAAAUGCAAACAUGUCUGGCAAACACAAAUCCCAUAAAUGUUU_UGUGC____CCC ......................................((((((((((.((((...((((.......))))....)))).))).......)))))))....................... (-13.16 = -13.16 + 0.00)
| Location | 6,260,944 – 6,261,034 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
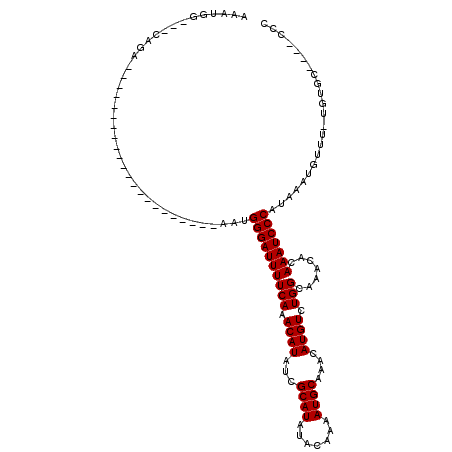
>2L_DroMel_CAF1 6260944 90 - 22407834 GGG----GCACA-AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCGUU----------------------UCUG---CCAUUU ..(----(((.(-(.......(((((((((((....))((((((((((.((((.......)))))))))))))).))))))))))----------------------).))---)).... ( -26.71) >DroVir_CAF1 122262 91 - 1 CAGCACCACACA-AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCAUU----------------------UC------CAUUG ............-........(((((((((((....))((((((((((.((((.......)))))))))))))).))))))))).----------------------..------..... ( -22.80) >DroPse_CAF1 172378 115 - 1 GGG----GCACA-AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCCUCCGUUCCACUCGUUUUGUGUGUUUCUGCUGCCAUUU ..(----(((((-(((((.....(((((((((....))((((((((((.((((.......)))))))))))))).)))))))........(((.......)))))))).)).)))).... ( -31.80) >DroGri_CAF1 105118 90 - 1 GGG----ACUCA-AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCAUU----------------------UCUG---CCACUG ...----.....-...((...(((((((((((....))((((((((((.((((.......)))))))))))))).))))))))).----------------------..))---...... ( -23.30) >DroMoj_CAF1 154596 95 - 1 CAGACACACACACAGACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCAUU----------------------UCUC---CCAUUG .....................(((((((((((....))((((((((((.((((.......)))))))))))))).))))))))).----------------------....---...... ( -22.80) >DroPer_CAF1 172835 115 - 1 GGG----GCACA-AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCCUCCGUUCCCCUCGUUUUGUGUGUUUCUGCUGCCAUUU .((----(((((-((((......(((((((((....))((((((((((.((((.......)))))))))))))).)))))))....(.......))))))))))((....))..)).... ( -30.80) >consensus GGG____ACACA_AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCAUU______________________UCUG___CCAUUG .....................(((((((((((....))((((((((((.((((.......)))))))))))))).))))))))).................................... (-21.42 = -21.67 + 0.25)


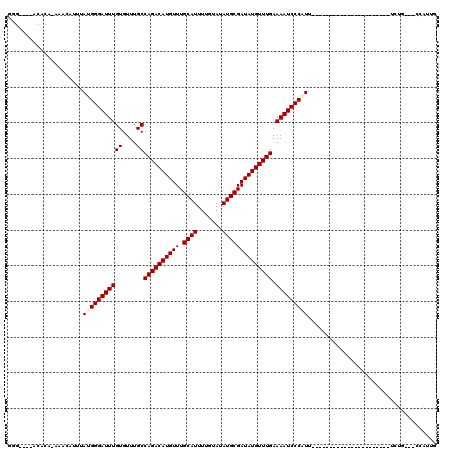
| Location | 6,260,959 – 6,261,065 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -19.94 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6260959 106 - 22407834 CCUCUAACUUCCCUUCGUGG---UUGUUGGGUGGGGGGCACAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC ((((((((..((......))---..)))))).)).(((((((((.(........).)))))))))..((((((((((.((((.......))))))))))))))...... ( -29.30) >DroPse_CAF1 172418 96 - 1 CCGCCCAUUC-------------CCAUUGAGAGAGGGGCACAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC ..((.....(-------------((.((....)))))(((((((.(........).)))))))..))((((((((((.((((.......))))))))))))))...... ( -25.00) >DroGri_CAF1 105133 90 - 1 -----------------AGGGAG--GCAGGUUGGGGGACUCAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC -----------------..(((.--(((((((((....)).....((....)).))))))).)))..((((((((((.((((.......))))))))))))))...... ( -21.10) >DroYak_CAF1 109644 97 - 1 CCUCCAACUUCCCUUCCAGG------------GGGGGGCACAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC .......(((((((....))------------)))))(((((((.(........).)))))))....((((((((((.((((.......))))))))))))))...... ( -28.60) >DroAna_CAF1 56891 92 - 1 CCCCUAACACCC-----ACG------------AUGGGGCACAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC .........(((-----...------------..)))(((((((.(........).)))))))....((((((((((.((((.......))))))))))))))...... ( -23.60) >DroPer_CAF1 172875 95 - 1 CCGCCCAUUC--------------CAUUGAGAGAGGGGCACAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC ...(((.(((--------------......))).)))(((((((.(........).)))))))....((((((((((.((((.......))))))))))))))...... ( -24.60) >consensus CCGCCAACUC_______AGG______UUG_G_GGGGGGCACAAAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUC ...................................(((((((((.(........).)))))))))..((((((((((.((((.......))))))))))))))...... (-19.94 = -19.97 + 0.03)

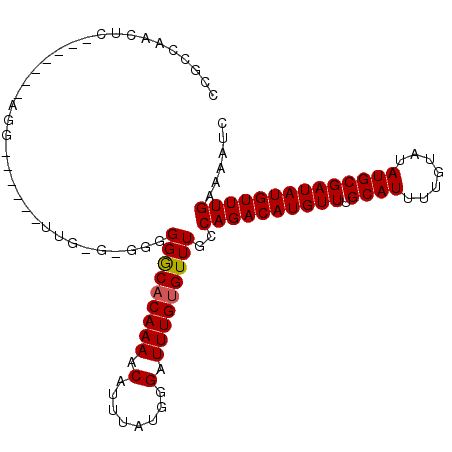
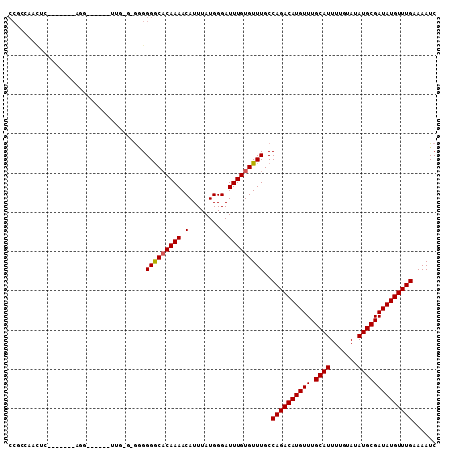
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:41 2006