
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
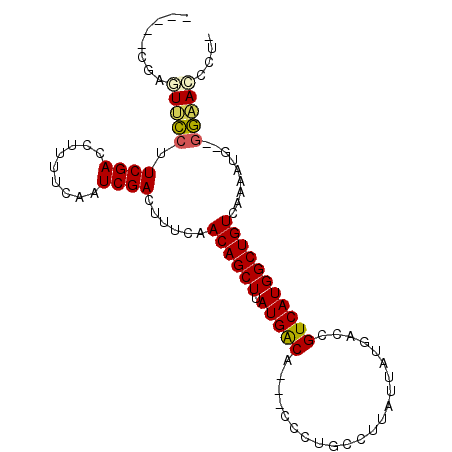
| Location | 6,250,156 – 6,250,250 |
| Length | 94 |
| Max. P | 0.745385 |

| Location | 6,250,156 – 6,250,250 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -15.38 |
| Energy contribution | -14.97 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
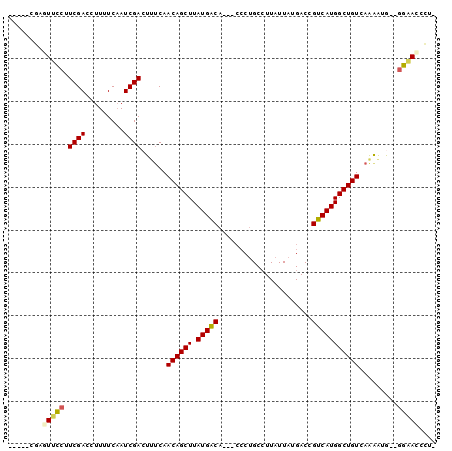
>2L_DroMel_CAF1 6250156 94 + 22407834 CGCUGCGAGUUCCUUCGACCUUUUCAAUCGACUUUCAACAGCUUAUGACA---CCCAGCCUUAUUAUGACCGUCAUGGCUGUCAAGAUG--GGAACCCG- .....((.((((((((((.........))))(((...((((((.(((((.---..((.........))...))))))))))).)))..)--))))).))- ( -21.70) >DroPse_CAF1 153196 94 + 1 -----CUUUUGUCUUCGACCUUUUCAAUCGACUCUCAACAGCUUAUGACAGGCCCCUGCCUUAUUAUGACUGUCAUGGCUGUCAGAGAAGAGAUACCCU- -----....(((((((((.........))))((((..((((((.(((((((..................))))))))))))).))))...)))))....- ( -24.47) >DroEre_CAF1 85056 94 + 1 CGCUGCGAGUUCCUUCGACCUUUUCCAUCGACUUUCAACAGCUUAUGACA---CCCUGCCUUAUUAUGACCGUCAUGGCUGUCAAAAUG--GGAACCCU- ......(.((((((((((.........)))).(((..((((((.(((((.---..................)))))))))))..))).)--))))).).- ( -19.71) >DroYak_CAF1 98620 94 + 1 CGCUCCGAGUUCCUUCGACCUUUUCAAUCGACUUUCAACAGCUUAUGACA---CCCUGCCUUAUUAUGACCGUCAUGGCUGUCAAAAUG--GGAACCCU- ......(.((((((((((.........)))).(((..((((((.(((((.---..................)))))))))))..))).)--))))).).- ( -19.71) >DroAna_CAF1 49009 89 + 1 -----GUCUUUCCAUCGACCUUUUCAAUCGACUACCAACAGCUUAUGGCA---GGCUGC-UUAUUAUGACUGUCAUGGCUGUCAAAAGG--AGCAACGUU -----((..((((.((((.........))))......((((((.((((((---(.....-.........))))))))))))).....))--))..))... ( -22.64) >DroPer_CAF1 152437 92 + 1 -----CUUUUGUCUUCGACCUUUUCAAUCGACUCUCAACAGCUUAUGACAGGCCCCUGCCUUAUUAUGACUGUCAUGGCUGUCAGAGAA--GAUACCCU- -----....(((((((((.........))))((((..((((((.(((((((..................))))))))))))).)))).)--))))....- ( -24.37) >consensus _____CGAGUUCCUUCGACCUUUUCAAUCGACUUUCAACAGCUUAUGACA___CCCUGCCUUAUUAUGACCGUCAUGGCUGUCAAAAUG__GGAACCCU_ ........(((((.((((.........))))......((((((.(((((......................))))))))))).........))))).... (-15.38 = -14.97 + -0.42)

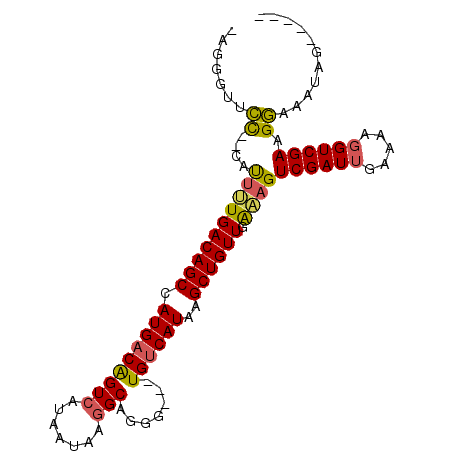
| Location | 6,250,156 – 6,250,250 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -16.17 |
| Energy contribution | -15.48 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
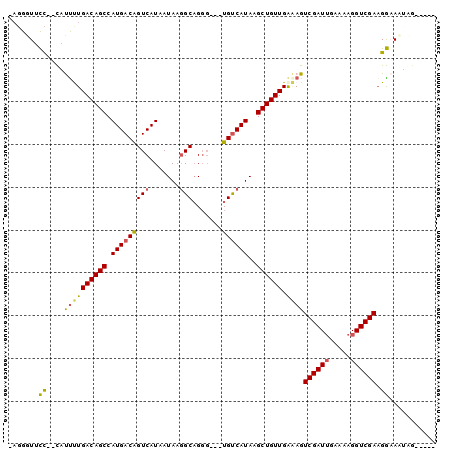
>2L_DroMel_CAF1 6250156 94 - 22407834 -CGGGUUCC--CAUCUUGACAGCCAUGACGGUCAUAAUAAGGCUGGG---UGUCAUAAGCUGUUGAAAGUCGAUUGAAAAGGUCGAAGGAACUCGCAGCG -((((((((--....(..(((((.((((((.(((.........))).---))))))..)))))..)...((((((.....)))))).))))))))..... ( -33.50) >DroPse_CAF1 153196 94 - 1 -AGGGUAUCUCUUCUCUGACAGCCAUGACAGUCAUAAUAAGGCAGGGGCCUGUCAUAAGCUGUUGAGAGUCGAUUGAAAAGGUCGAAGACAAAAG----- -......(((.(((((.((((((.(((.....))).....(((((....)))))....)))))))))))((((((.....)))))))))......----- ( -25.30) >DroEre_CAF1 85056 94 - 1 -AGGGUUCC--CAUUUUGACAGCCAUGACGGUCAUAAUAAGGCAGGG---UGUCAUAAGCUGUUGAAAGUCGAUGGAAAAGGUCGAAGGAACUCGCAGCG -.(((((((--..(((..(((((.((((((.((...........)).---))))))..)))))..))).(((((.......))))).)))))))...... ( -32.20) >DroYak_CAF1 98620 94 - 1 -AGGGUUCC--CAUUUUGACAGCCAUGACGGUCAUAAUAAGGCAGGG---UGUCAUAAGCUGUUGAAAGUCGAUUGAAAAGGUCGAAGGAACUCGGAGCG -.(((((((--..(((..(((((.((((((.((...........)).---))))))..)))))..))).((((((.....)))))).)))))))...... ( -33.50) >DroAna_CAF1 49009 89 - 1 AACGUUGCU--CCUUUUGACAGCCAUGACAGUCAUAAUAA-GCAGCC---UGCCAUAAGCUGUUGGUAGUCGAUUGAAAAGGUCGAUGGAAAGAC----- ...((((((--..((.((((..........)))).))..)-)))))(---(((((........))))))((.(((((.....))))).)).....----- ( -23.10) >DroPer_CAF1 152437 92 - 1 -AGGGUAUC--UUCUCUGACAGCCAUGACAGUCAUAAUAAGGCAGGGGCCUGUCAUAAGCUGUUGAGAGUCGAUUGAAAAGGUCGAAGACAAAAG----- -........--(((((.((((((.(((.....))).....(((((....)))))....)))))))))))((((((.....)))))).........----- ( -25.20) >consensus _AGGGUUCC__CAUUUUGACAGCCAUGACAGUCAUAAUAAGGCAGGG___UGUCAUAAGCUGUUGAAAGUCGAUUGAAAAGGUCGAAGGAAAUAG_____ .......((....((((((((((.(((((((((.......))).......))))))..)))))).))))((((((.....)))))).))........... (-16.17 = -15.48 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:36 2006