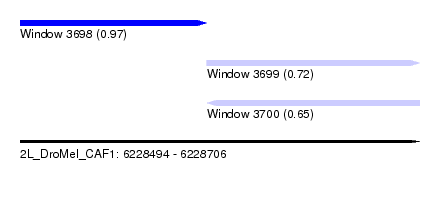
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,228,494 – 6,228,706 |
| Length | 212 |
| Max. P | 0.969207 |

| Location | 6,228,494 – 6,228,593 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -20.83 |
| Energy contribution | -20.79 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6228494 99 + 22407834 CACAACAAGCUGCUUUCCCACGCAGCGGAAAACUUUGAUGAGCCUCAUUGAAAUUGAAUUUCCUACCAAGUGCCUUAC---------------------CGUGCAAUUUGUGUUUUCCCA ........(((((........)))))(((((((((..(((.....)))..))..............((((((((....---------------------.).)).))))).))))))).. ( -23.80) >DroSec_CAF1 64999 99 + 1 CACAACAAGCUGCUUUCCCACGCAGCGGAAAACUUUGAUGAGCAUCAUUGAAAUUGAAUUUCCUACCAAGUGCCUCAC---------------------CGUGCAAUUUGUGUUUUCCCA ........(((((........)))))(((((((((..(((.....)))..))..............((((((((....---------------------.).)).))))).))))))).. ( -23.80) >DroSim_CAF1 78182 99 + 1 CACAACAAGCUGCUUUCCCACGCAGCGGAAAACUUUGAUGAGGCUCAUUGAAAUUGAAUUUCCUACCAAGUGCCUCAC---------------------CGUGCAAUUUGUGUUUUCCCA ........(((((........)))))(((((((...(.((((((...(((................)))..)))))).---------------------)..((.....))))))))).. ( -26.29) >DroEre_CAF1 62093 98 + 1 CACAACAAGCCGCUUUCCCACGCGGCGGAAAACUUUGAUGAGCCUCAUUGAAAUUGAAUUUCCUUCCAAGUGC-UCAC---------------------CGUGCAAUUUGUGUUUUCCCA ........(((((........)))))(((((((((..(((.....)))..))..............(((((((-....---------------------...)).))))).))))))).. ( -26.10) >DroYak_CAF1 76653 119 + 1 CACAACAAGCCGCUUUCCCACGCAGCGGAAAACUUUGAUGAGCCUCAUUGAAAUUGAAUUUCCUUCCAAGUGC-UCACCGCUCACCGUUCUCCGUUCUCCGUGCAAUUUGUGUUUUCCCA (((((...(((((........))((((((.(((..((((((((((....(((((...)))))......)).))-)))....)))..))).))))))....).))...)))))........ ( -23.80) >consensus CACAACAAGCUGCUUUCCCACGCAGCGGAAAACUUUGAUGAGCCUCAUUGAAAUUGAAUUUCCUACCAAGUGCCUCAC_____________________CGUGCAAUUUGUGUUUUCCCA ........(((((........)))))(((((((((..(((.....)))..))..............(((((((.............................)).))))).))))))).. (-20.83 = -20.79 + -0.04)

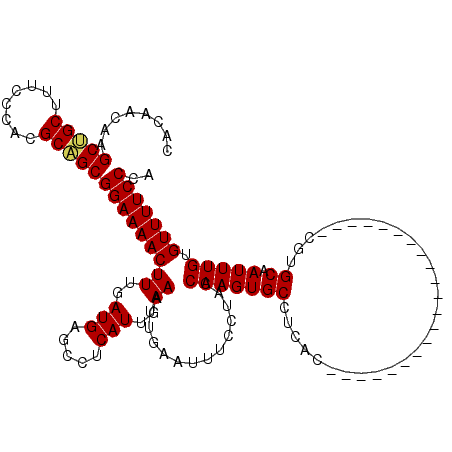
| Location | 6,228,593 – 6,228,706 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -29.15 |
| Energy contribution | -29.07 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
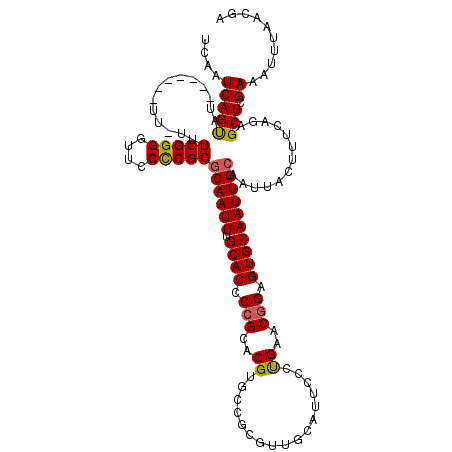
>2L_DroMel_CAF1 6228593 113 + 22407834 UCGUUAAAUUUCGAGCUCUGAAAGUAAUUGCAAUUGCACUCCGUUCAGGGAAUGCAACGCGGCACGUGCGGGGUGCAAAUUGCCCGGGGAACCCCGAAAAAA-------AUACUGAUUGA (((........)))..((....((((...(((((((((((((((...(....(((...)))...)..))))))))).)))))).((((....))))......-------.))))....)) ( -34.80) >DroSec_CAF1 65098 112 + 1 UCGUUAAAUUUCGAGCUCUGAAAGUAAUUGCAAUUGCACUCCGUUCAGGGAAUGCAACGCGGCACGUGCGGGGUGCAAAUUGCCCGGGGAACCCCGAAA-AA-------AUACUGAUUGA (((........)))..((....((((...(((((((((((((((...(....(((...)))...)..))))))))).)))))).((((....))))...-..-------.))))....)) ( -34.80) >DroSim_CAF1 78281 112 + 1 UCGUUAAAUUUCGAGCUCUGAAAGUAAUUGCAAUUGCACUCCGUUCAGGGAAUGCAACGCGGCACGUGCGGGGUGCAAAUUGCCCGGGGAACCCCGAAA-AA-------AUACUGAUUGA (((........)))..((....((((...(((((((((((((((...(....(((...)))...)..))))))))).)))))).((((....))))...-..-------.))))....)) ( -34.80) >DroEre_CAF1 62191 98 + 1 UCGUUAAACUUCGAGCUCUGAAAGUAAUUGCAAUUGCACUCAGUUCG-------------GGCACGUGCGGGGUGCAAAUUGCCCGAGGAACCCCGAAA-AA--------UGCUGAUUGA (((.....(((((.(((.....)))....((((((((((((.((.((-------------....)).)).)))))).)))))).))))).....)))..-..--------.......... ( -25.60) >DroYak_CAF1 76772 107 + 1 UCGUUAAAUUUCGAGCUCUGAAAGUAAUUGCAAUUGCACUCCGCUCGG------------GGCACGUGCGGGGUGCAAAUUGUCCGGGGAACCCCGAAA-AAAAAUAGUAUACUGAUUGA (((........)))..((....((((...(((((((((((((((.((.------------....)).))))))))).)))))).((((....))))...-..........))))....)) ( -36.60) >consensus UCGUUAAAUUUCGAGCUCUGAAAGUAAUUGCAAUUGCACUCCGUUCAGGGAAUGCAACGCGGCACGUGCGGGGUGCAAAUUGCCCGGGGAACCCCGAAA_AA_______AUACUGAUUGA (((........)))..((....((((...(((((((((((((((.(...................).))))))))).)))))).((((....))))..............))))....)) (-29.15 = -29.07 + -0.08)

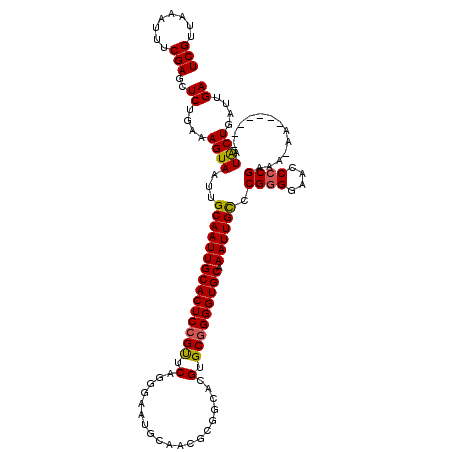
| Location | 6,228,593 – 6,228,706 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.09 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6228593 113 - 22407834 UCAAUCAGUAU-------UUUUUUCGGGGUUCCCCGGGCAAUUUGCACCCCGCACGUGCCGCGUUGCAUUCCCUGAACGGAGUGCAAUUGCAAUUACUUUCAGAGCUCGAAAUUUAACGA ....(((((..-------......((((....))))((((...(((.....)))..))))((((((((((((......))))))))).))).............))).)).......... ( -34.50) >DroSec_CAF1 65098 112 - 1 UCAAUCAGUAU-------UU-UUUCGGGGUUCCCCGGGCAAUUUGCACCCCGCACGUGCCGCGUUGCAUUCCCUGAACGGAGUGCAAUUGCAAUUACUUUCAGAGCUCGAAAUUUAACGA .......((..-------..-(((((((.(((....((((...(((.....)))..))))((((((((((((......))))))))).)))...........))))))))))....)).. ( -34.80) >DroSim_CAF1 78281 112 - 1 UCAAUCAGUAU-------UU-UUUCGGGGUUCCCCGGGCAAUUUGCACCCCGCACGUGCCGCGUUGCAUUCCCUGAACGGAGUGCAAUUGCAAUUACUUUCAGAGCUCGAAAUUUAACGA .......((..-------..-(((((((.(((....((((...(((.....)))..))))((((((((((((......))))))))).)))...........))))))))))....)).. ( -34.80) >DroEre_CAF1 62191 98 - 1 UCAAUCAGCA--------UU-UUUCGGGGUUCCUCGGGCAAUUUGCACCCCGCACGUGCC-------------CGAACUGAGUGCAAUUGCAAUUACUUUCAGAGCUCGAAGUUUAACGA ..........--------..-(((((((.....(((((((...(((.....)))..))))-------------))).((((((((....)))......)))))..)))))))........ ( -28.30) >DroYak_CAF1 76772 107 - 1 UCAAUCAGUAUACUAUUUUU-UUUCGGGGUUCCCCGGACAAUUUGCACCCCGCACGUGCC------------CCGAGCGGAGUGCAAUUGCAAUUACUUUCAGAGCUCGAAAUUUAACGA ....(((((...........-.((((((....))))))(((((.((((.((((.((....------------.)).)))).)))))))))..............))).)).......... ( -29.20) >consensus UCAAUCAGUAU_______UU_UUUCGGGGUUCCCCGGGCAAUUUGCACCCCGCACGUGCCGCGUUGCAUUCCCUGAACGGAGUGCAAUUGCAAUUACUUUCAGAGCUCGAAAUUUAACGA ....(((((..............(((((....)))))((((((.((((.(((..((.................))..))).)))))))))).............))).)).......... (-23.25 = -23.09 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:33 2006