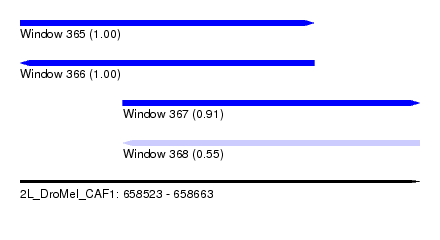
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 658,523 – 658,663 |
| Length | 140 |
| Max. P | 0.999936 |

| Location | 658,523 – 658,626 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -40.06 |
| Consensus MFE | -35.77 |
| Energy contribution | -35.93 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
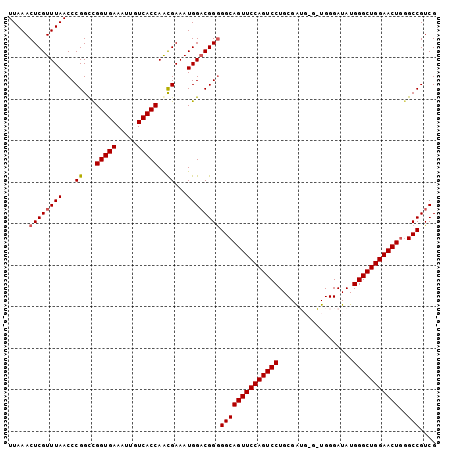
>2L_DroMel_CAF1 658523 103 + 22407834 UUAAACUCGUUUAACCCGGCCGGUGAAAUUGUCACCAGCGAAAUGGACGGAGGCAGUUCCAGUCCUGCGAUGCGAUGGGAUAUGGGCUGGAACUGGGCCGUCG ..................((.(((((.....))))).))......(((((...((((((((((((....((.(....).))..))))))))))))..))))). ( -40.90) >DroSec_CAF1 47138 103 + 1 UUAAACUCGUUUAACCCGGCCGGUGAAAUUGUCACCAACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUGGGGUGGGAUAUGGGCUGGAACUGGGCCGUCG .....((((((((...((...(((((.....)))))..))...))))))))((((((((((((((..................)))))))))))..))).... ( -38.37) >DroSim_CAF1 51669 103 + 1 UUAAACUCGUUUAACCCGGCCGGUGAAAUUGUCACCAACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUGGGAUGGGAUAUGGGCUGGAACUGUGCCGUCG .......((((((...((...(((((.....)))))..))...))))))((.(((((((((((((..................))))))))))))).)).... ( -39.87) >DroEre_CAF1 52090 98 + 1 UUAAACUCGUUUAACCCGGUUGGUGAAAUUGUCACCAAUGAAAUGGACGGGGGCAGUUCCAGUCCUGCGA-----UGGGAUAUGGGCUGGAACUUGGCCGUCG .....((((((((.....((((((((.....))))))))....))))))))((((((((((((((.....-----........)))))))))))..))).... ( -40.32) >DroYak_CAF1 51496 98 + 1 UUAAACUCGUUUAACCCGGUCGGUGAAAUUGUCACCAAUGAAAUGGCCGGGGGCAGUUCCAGUCCUGCGG-----UGGGAUAUGGGCUGGAACUGGGCCGUCG ..............((((((((((((.....)))))........)))))))((((((((((((((.....-----........)))))))))))..))).... ( -40.82) >consensus UUAAACUCGUUUAACCCGGCCGGUGAAAUUGUCACCAACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUG_G_UGGGAUAUGGGCUGGAACUGGGCCGUCG .....((((((((...((...(((((.....)))))..))...))))))))((((((((((((((..................)))))))))))..))).... (-35.77 = -35.93 + 0.16)

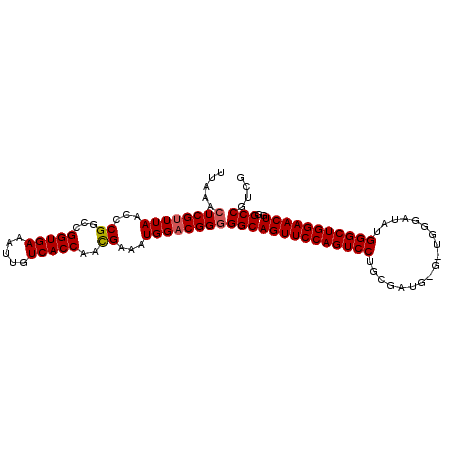
| Location | 658,523 – 658,626 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -31.65 |
| Energy contribution | -31.89 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 658523 103 - 22407834 CGACGGCCCAGUUCCAGCCCAUAUCCCAUCGCAUCGCAGGACUGGAACUGCCUCCGUCCAUUUCGCUGGUGACAAUUUCACCGGCCGGGUUAAACGAGUUUAA .(((((..(((((((((.((..................)).)))))))))...))))).(((..((((((((.....))))))))..)))............. ( -36.47) >DroSec_CAF1 47138 103 - 1 CGACGGCCCAGUUCCAGCCCAUAUCCCACCCCAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUUGGUGACAAUUUCACCGGCCGGGUUAAACGAGUUUAA .(((((..(((((((((.((..................)).)))))))))...))))).(((..((((((((.....))))))))..)))............. ( -33.97) >DroSim_CAF1 51669 103 - 1 CGACGGCACAGUUCCAGCCCAUAUCCCAUCCCAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUUGGUGACAAUUUCACCGGCCGGGUUAAACGAGUUUAA .(((((..(((((((((.((..................)).)))))))))...))))).(((..((((((((.....))))))))..)))............. ( -33.97) >DroEre_CAF1 52090 98 - 1 CGACGGCCAAGUUCCAGCCCAUAUCCCA-----UCGCAGGACUGGAACUGCCCCCGUCCAUUUCAUUGGUGACAAUUUCACCAACCGGGUUAAACGAGUUUAA .(((((...((((((((.((........-----.....)).))))))))....))))).......(((((((.....))))))).((.......))....... ( -30.02) >DroYak_CAF1 51496 98 - 1 CGACGGCCCAGUUCCAGCCCAUAUCCCA-----CCGCAGGACUGGAACUGCCCCCGGCCAUUUCAUUGGUGACAAUUUCACCGACCGGGUUAAACGAGUUUAA .((((((..((((((((.((........-----.....)).)))))))))))(((((........(((((((.....))))))))))))........)))... ( -33.22) >consensus CGACGGCCCAGUUCCAGCCCAUAUCCCA_C_CAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUUGGUGACAAUUUCACCGGCCGGGUUAAACGAGUUUAA .(((((..(((((((((.((..................)).)))))))))...))))).((((.((((((((.....)))))))).))))............. (-31.65 = -31.89 + 0.24)

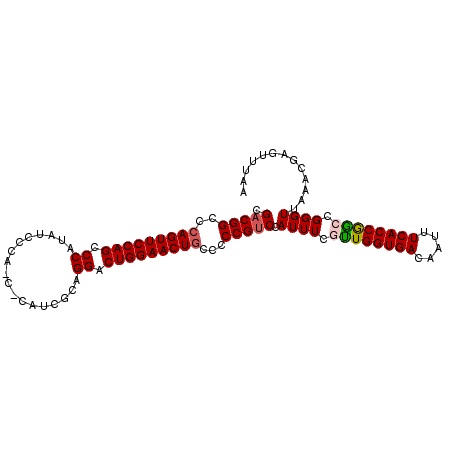
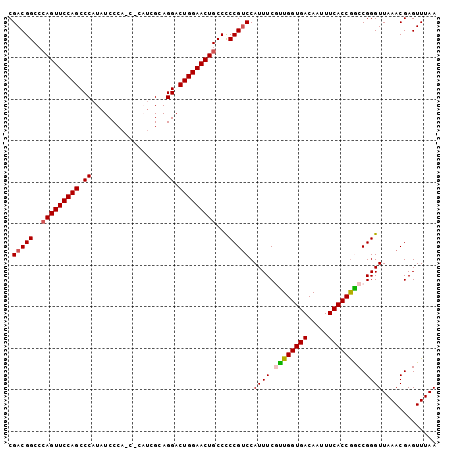
| Location | 658,559 – 658,663 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -21.50 |
| Energy contribution | -23.75 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 658559 104 + 22407834 AGCGAAAUGGACGGAGGCAGUUCCAGUCCUGCGAUGCGAUGGGAUAUG---GGCUGGAACUGGGCCGUCGUCAGUCCAGAGAUUGGCAAUUUUAUCUGUCAACAACA ...(((((.(((((...((((((((((((....((.(....).))..)---)))))))))))..)))))(((((((....))))))).))))).............. ( -40.00) >DroSec_CAF1 47174 104 + 1 AACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUGGGGUGGGAUAUG---GGCUGGAACUGGGCCGUCGUCAGUCCAGAGAUUGGCAAUUUUAUCUGUCAACAACA ...(((((.(((((...((((((((((((..................)---)))))))))))..)))))(((((((....))))))).))))).............. ( -37.57) >DroSim_CAF1 51705 104 + 1 AACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGAUGGGAUGGGAUAUG---GGCUGGAACUGUGCCGUCGUCAGUCCAGAGAUUGGCAAUUUUAUCUGUCAACAACA ...(((((.(((((.(.((((((((((((..................)---)))))))))))).)))))(((((((....))))))).))))).............. ( -39.57) >DroEre_CAF1 52126 99 + 1 AAUGAAAUGGACGGGGGCAGUUCCAGUCCUGCGA-----UGGGAUAUG---GGCUGGAACUUGGCCGUCGUCAGUCCAGAGAUUGGGAAUUUUAUCUGUCAACAACA .(((((((.(((((.((((((((((((((.....-----........)---))))))))))..))).)))))..(((........))))))))))............ ( -36.32) >DroWil_CAF1 18009 85 + 1 AGAGUGUCGGUCGAAGACU-----------GG-------CAGGGCAUGAACGGCUGAA---GGGCUAUGG-CCGCCAGAAGACAGCCAACUUUAUCUGUCAACAACA ....((((((((...))))-----------))-------)).((((((((.(((((..---.(((.....-..)))......)))))...))))..))))....... ( -23.10) >DroYak_CAF1 51532 99 + 1 AAUGAAAUGGCCGGGGGCAGUUCCAGUCCUGCGG-----UGGGAUAUG---GGCUGGAACUGGGCCGUCGUCAGUCCAGAGAUUGGCAAUUUUAUCUGUCAACAACU .(((((((((((.....((((((((((((.....-----........)---)))))))))))))))...(((((((....))))))).)))))))............ ( -37.42) >consensus AACGAAAUGGACGGGGGCAGUUCCAGUCCUGCGA_____UGGGAUAUG___GGCUGGAACUGGGCCGUCGUCAGUCCAGAGAUUGGCAAUUUUAUCUGUCAACAACA .........(((((((((((((((((((.......................))))))))))..)))...(((((((....))))))).......))))))....... (-21.50 = -23.75 + 2.25)

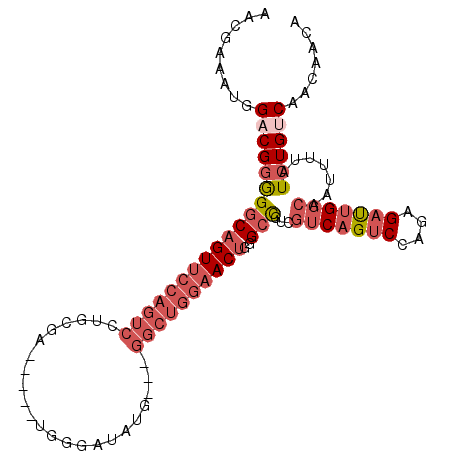
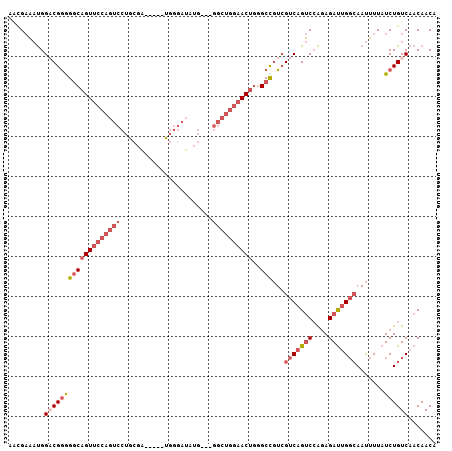
| Location | 658,559 – 658,663 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -15.99 |
| Energy contribution | -18.02 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 658559 104 - 22407834 UGUUGUUGACAGAUAAAAUUGCCAAUCUCUGGACUGACGACGGCCCAGUUCCAGCC---CAUAUCCCAUCGCAUCGCAGGACUGGAACUGCCUCCGUCCAUUUCGCU .((((((.(.((((..........)))).).))).)))(((((..(((((((((.(---(..................)).)))))))))...)))))......... ( -28.17) >DroSec_CAF1 47174 104 - 1 UGUUGUUGACAGAUAAAAUUGCCAAUCUCUGGACUGACGACGGCCCAGUUCCAGCC---CAUAUCCCACCCCAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUU .((((((.(.((((..........)))).).))).)))(((((..(((((((((.(---(..................)).)))))))))...)))))......... ( -28.17) >DroSim_CAF1 51705 104 - 1 UGUUGUUGACAGAUAAAAUUGCCAAUCUCUGGACUGACGACGGCACAGUUCCAGCC---CAUAUCCCAUCCCAUCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUU .((((((.(.((((..........)))).).))).)))(((((..(((((((((.(---(..................)).)))))))))...)))))......... ( -28.17) >DroEre_CAF1 52126 99 - 1 UGUUGUUGACAGAUAAAAUUCCCAAUCUCUGGACUGACGACGGCCAAGUUCCAGCC---CAUAUCCCA-----UCGCAGGACUGGAACUGCCCCCGUCCAUUUCAUU .((((((.(.((((..........)))).).))).)))(((((...((((((((.(---(........-----.....)).))))))))....)))))......... ( -26.42) >DroWil_CAF1 18009 85 - 1 UGUUGUUGACAGAUAAAGUUGGCUGUCUUCUGGCGG-CCAUAGCCC---UUCAGCCGUUCAUGCCCUG-------CC-----------AGUCUUCGACCGACACUCU (((((((((.((((.....((((((((....)))))-)))..((..---....)).............-------..-----------.)))))))).))))).... ( -22.40) >DroYak_CAF1 51532 99 - 1 AGUUGUUGACAGAUAAAAUUGCCAAUCUCUGGACUGACGACGGCCCAGUUCCAGCC---CAUAUCCCA-----CCGCAGGACUGGAACUGCCCCCGGCCAUUUCAUU .(((((((.((((..............))))...)))))))(((((((((((((.(---(........-----.....)).))))))))).....))))........ ( -30.56) >consensus UGUUGUUGACAGAUAAAAUUGCCAAUCUCUGGACUGACGACGGCCCAGUUCCAGCC___CAUAUCCCA_____UCGCAGGACUGGAACUGCCCCCGUCCAUUUCGUU .((((((.(.((((..........)))).).))).)))(((((..(((((((((((......................)).)))))))))...)))))......... (-15.99 = -18.02 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:38 2006