
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,217,635 – 6,217,739 |
| Length | 104 |
| Max. P | 0.856905 |

| Location | 6,217,635 – 6,217,739 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -13.32 |
| Energy contribution | -14.60 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
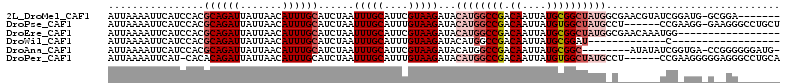
>2L_DroMel_CAF1 6217635 104 + 22407834 AUUAAAAUUCAUCCACGCAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUCGUAAGAUACAUGGCCGACAAUUAUGCGGCUAUGGCGAACGUAUCGGAUG-GCGGA------- ........((((((..((((((.......))))))......(((((..((....))..((((((((.((....)))))))))))))))......)))))-)....------- ( -32.00) >DroPse_CAF1 88878 105 + 1 AUUAAAAUUCAUCCACGCAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUUGUAAGAUACAUGGCCGACAAUUAUGUGGCUAUGCCU------CCGAAGG-GAAGGGCCUGCU ................((((.......(((..((((.......))))..)))......(((((((.(((....))))))))))(((------((....)-).)))..)))). ( -31.00) >DroEre_CAF1 53002 95 + 1 AUUAAAAUUCAUCCACGCAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUCGUAAGAUACAUGGCCGACAAUUAUGCGGCUAUGGCGAACAAAUGG----------------- ............(((.((((((.......))))))......(((((..((....))..((((((((.((....)))))))))))))))....)))----------------- ( -25.20) >DroWil_CAF1 46326 81 + 1 AUUAAAAUUCAUCCACGCAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUUGUAAGAUACAUGGCCGACAAUUAUGCGGAU-------------C------------------ ..........((((..((((((.......)))))).........((((((((..(........)..))))..))))))))-------------.------------------ ( -13.30) >DroAna_CAF1 25422 102 + 1 AUUAAAAUUCAUCCACGCAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUCGUAAGAUACAUGGCCGACAAUUAUGCGGC--------AUAUAUCGGUGA-CCGGGGGGAUG- .........(((((..((((((((..............)))))))).((((...((((.(((.(((.((....))))))--------)).))))(....-))))).)))))- ( -24.34) >DroPer_CAF1 89013 105 + 1 AUUAAAAUUCAU-CACACAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUUGUAAGAUACAUGGCCGACAAUUAUGUGGCUAUGCCU------CCGAAGGGGGAGGGCCUGCA ............-....(((.......(((..((((.......))))..)))......(((((((.(((....))))))))))(((------((......)))))..))).. ( -28.50) >consensus AUUAAAAUUCAUCCACGCAGAUUAUUAACAUUUGCAUCUAAUUUGCAUUCGUAAGAUACAUGGCCGACAAUUAUGCGGCUAUG_C_______UCG_AGG_G__GG_______ ................((((((.......))))))......(((((....)))))...((((((((.((....))))))))))............................. (-13.32 = -14.60 + 1.28)
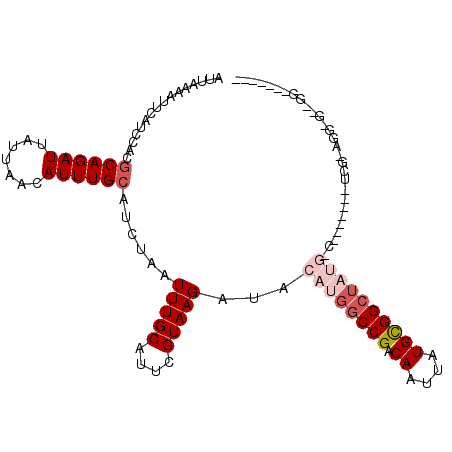
| Location | 6,217,635 – 6,217,739 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.31 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6217635 104 - 22407834 -------UCCGC-CAUCCGAUACGUUCGCCAUAGCCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAAU -------.....-((((((((((((........((((((....)).)))).)))))))......(((((.((((...((....))...)))).))))))))))......... ( -26.40) >DroPse_CAF1 88878 105 - 1 AGCAGGCCCUUC-CCUUCGG------AGGCAUAGCCACAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAAU .(((((((..((-(....))------))))...((((((....))).)))..(((((((..............)))))))............))))................ ( -23.54) >DroEre_CAF1 53002 95 - 1 -----------------CCAUUUGUUCGCCAUAGCCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAAU -----------------..(((..((((((((.((((((....)).)))).)))............(((.((((...((....))...)))).))))))))..)))...... ( -22.80) >DroWil_CAF1 46326 81 - 1 ------------------G-------------AUCCGCAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAAU ------------------.-------------((((((.(((((((..((..(((((((..............)))))))...))..))))).)).)))))).......... ( -17.84) >DroAna_CAF1 25422 102 - 1 -CAUCCCCCCGG-UCACCGAUAUAU--------GCCGCAUAAUUGUCGGCCAUGUAUCUUACGAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAAU -(((((......-.....(((((((--------((((((....)).))).))))))))......(((((.((((...((....))...)))).))))))))))......... ( -28.10) >DroPer_CAF1 89013 105 - 1 UGCAGGCCCUCCCCCUUCGG------AGGCAUAGCCACAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGUGUG-AUGAAUUUUAAU .((..(((.(((......))------))))...))(((((((((((..((..(((((((..............)))))))...))..))))).))))))-............ ( -26.04) >consensus _______CC__C_CCU_CGA_______G_CAUAGCCGCAUAAUUGUCGGCCAUGUAUCUUACAAAUGCAAAUUAGAUGCAAAUGUUAAUAAUCUGCGUGGAUGAAUUUUAAU ..................................((((.(((((((..((..(((((((..............)))))))...))..))))).)).))))............ (-14.36 = -14.31 + -0.06)

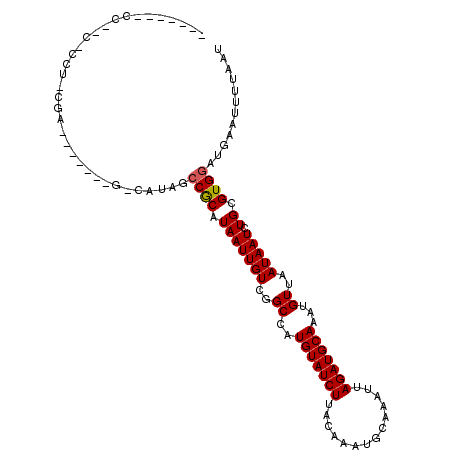
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:24 2006