| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,214,806 – 6,214,927 |
| Length | 121 |
| Max. P | 0.962131 |

| Location | 6,214,806 – 6,214,904 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.05 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -18.93 |
| Energy contribution | -20.65 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6214806 98 + 22407834 ------UCAU---UG-CCUGGUAUCUAGUUAAUGGGAUGCUUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG--- ------....---((-((.(((((((........)))))))......))))..((.((((((.((((((((((.(.......).)))))))))).)))))).))....--- ( -30.20) >DroPse_CAF1 83840 98 + 1 AGGGCUACAGAGGAGUCCU-GGAUCUAGCUAAUAGGAUGCUUAUGAUGAUACUUUGAUAAUGGCACAA---------CAGAACAGAACCAGCUGAUGUUGUGGAUUUG--- ((((((.......))))))-..(((.(((.........)))...)))................(((((---------((...(((......))).)))))))......--- ( -20.50) >DroSec_CAF1 51421 98 + 1 ------UCAC---AG-CCUGGUAUCUAGUUAAUGGGAUGCUUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG--- ------...(---((-(..(((((((........)))))))...)))).....((.((((((.((((((((((.(.......).)))))))))).)))))).))....--- ( -31.80) >DroSim_CAF1 62749 92 + 1 ------UCAC---UG-C------UCUAGUUAAUGGGAUGCUUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG--- ------((((---..-.------....((((.(..((((((......))))))..).))))..((((((((((.(.......).)))))))))).....)))).....--- ( -26.00) >DroEre_CAF1 50308 98 + 1 ------UCCC---GG-CCUGGUAUCUAGUUAAUGGGAUGCUUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAGGAGGAACUGCUGAUAUUGUGGAUUGG--- ------..((---((-(..(((((((........)))))))...)))))....((.((((((.((((((((((.(......)..)))))))))).)))))).))....--- ( -33.40) >DroYak_CAF1 62836 101 + 1 ------UCAC---AG-CCUGGUAUCUAGUUAAUGGGAUGCUUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUGGAAG ------...(---((-(..(((((((........)))))))...)))).....((.((((((.((((((((((.(.......).)))))))))).)))))).))....... ( -31.80) >consensus ______UCAC___AG_CCUGGUAUCUAGUUAAUGGGAUGCUUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG___ ......................((((......(((((((((......)))))))))((((((.((((((((((.(.......).)))))))))).))))))))))...... (-18.93 = -20.65 + 1.72)


| Location | 6,214,836 – 6,214,927 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.07 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -11.09 |
| Energy contribution | -13.53 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6214836 91 + 22407834 UUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG-----UGAAAAAA---------------AAACCACCGAUGGAA ....((((((......((((((.((((((((((.(.......).)))))))))).))))))((.(((.-----.......)---------------)).)))))))).... ( -26.90) >DroPse_CAF1 83879 97 + 1 UUAUGAUGAUACUUUGAUAAUGGCACAA---------CAGAACAGAACCAGCUGAUGUUGUGGAUUUG-----CGCCAGAAGGAAAAGACUUCACAGAAUGACAGAAGGCA .......................(((((---------((...(((......))).)))))))......-----.(((.((((.......)))).(.....)......))). ( -18.50) >DroSec_CAF1 51451 88 + 1 UUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG-----UGAAG------------------AAACCACCGAUGGGA ....((((((......((((((.((((((((((.(.......).)))))))))).))))))((.(((.-----....)------------------)).)))))))).... ( -26.70) >DroSim_CAF1 62773 88 + 1 UUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG-----UGAGG------------------AAACCACCGUUGGGC .............((.((((((.((((((((((.(.......).)))))))))).)))))).))...(-----((.(.------------------...))))........ ( -25.30) >DroYak_CAF1 62866 93 + 1 UUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUGGAAGAUUGAAA------------------GAACAACUGAUGGCA ....((((...((((.((((((.((((((((((.(.......).)))))))))).)))))).((((....))))))))------------------...))))........ ( -23.80) >DroPer_CAF1 84499 97 + 1 UUAUGAUGAUACUUUGAUAAUGGCACAA---------CAGAACAGAACCAGCUGAUGUUGUGGAUUUG-----CGCCAGAAGGAAAAGACUUCAUAGAAUGACAGAAGGCA ((((.((((..((((.....((((((((---------((...(((......))).))))))(......-----)))))......))))...))))...))))......... ( -18.10) >consensus UUAUGUUGGUAUUUUGAUAAUGGCAGCAGUUCUGCGUAAUAAGAGGAACUGCUGAUAUUGUGGAUUUG_____UGAAA__________________AAACCACCGAUGGCA .............((.((((((.((((((((((...........)))))))))).)))))).))............................................... (-11.09 = -13.53 + 2.45)

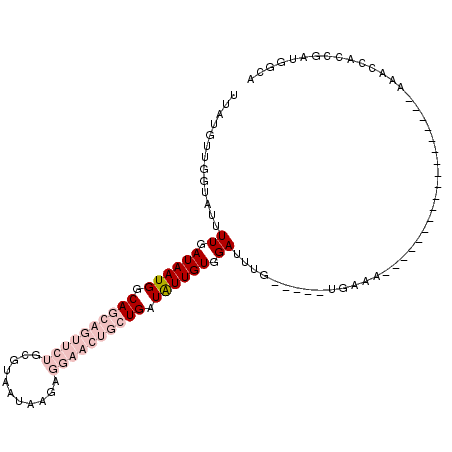
| Location | 6,214,836 – 6,214,927 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.07 |
| Mean single sequence MFE | -16.55 |
| Consensus MFE | -4.29 |
| Energy contribution | -6.62 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6214836 91 - 22407834 UUCCAUCGGUGGUUU---------------UUUUUUCA-----CAAAUCCACAAUAUCAGCAGUUCCUCUUAUUACGCAGAACUGCUGCCAUUAUCAAAAUACCAACAUAA ........((((.((---------------(.......-----.))).))))(((..(((((((((..(.......)..)))))))))..))).................. ( -16.90) >DroPse_CAF1 83879 97 - 1 UGCCUUCUGUCAUUCUGUGAAGUCUUUUCCUUCUGGCG-----CAAAUCCACAACAUCAGCUGGUUCUGUUCUG---------UUGUGCCAUUAUCAAAGUAUCAUCAUAA ...............(((((.(.((((......(((((-----(((....(((..(((....)))..)))....---------)))))))).....))))...).))))). ( -18.20) >DroSec_CAF1 51451 88 - 1 UCCCAUCGGUGGUUU------------------CUUCA-----CAAAUCCACAAUAUCAGCAGUUCCUCUUAUUACGCAGAACUGCUGCCAUUAUCAAAAUACCAACAUAA ........(((((((------------------.....-----.))).))))(((..(((((((((..(.......)..)))))))))..))).................. ( -16.50) >DroSim_CAF1 62773 88 - 1 GCCCAACGGUGGUUU------------------CCUCA-----CAAAUCCACAAUAUCAGCAGUUCCUCUUAUUACGCAGAACUGCUGCCAUUAUCAAAAUACCAACAUAA ........(((((((------------------.....-----.))).))))(((..(((((((((..(.......)..)))))))))..))).................. ( -16.50) >DroYak_CAF1 62866 93 - 1 UGCCAUCAGUUGUUC------------------UUUCAAUCUUCCAAUCCACAAUAUCAGCAGUUCCUCUUAUUACGCAGAACUGCUGCCAUUAUCAAAAUACCAACAUAA ........((((...------------------........................(((((((((..(.......)..)))))))))..(((.....)))..)))).... ( -13.00) >DroPer_CAF1 84499 97 - 1 UGCCUUCUGUCAUUCUAUGAAGUCUUUUCCUUCUGGCG-----CAAAUCCACAACAUCAGCUGGUUCUGUUCUG---------UUGUGCCAUUAUCAAAGUAUCAUCAUAA ...............(((((.(.((((......(((((-----(((....(((..(((....)))..)))....---------)))))))).....))))...).))))). ( -18.20) >consensus UGCCAUCGGUGGUUC__________________UUUCA_____CAAAUCCACAAUAUCAGCAGUUCCUCUUAUUACGCAGAACUGCUGCCAUUAUCAAAAUACCAACAUAA .........................................................(((((((((.............)))))))))....................... ( -4.29 = -6.62 + 2.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:21 2006