| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,186,336 – 6,186,450 |
| Length | 114 |
| Max. P | 0.988778 |

| Location | 6,186,336 – 6,186,450 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
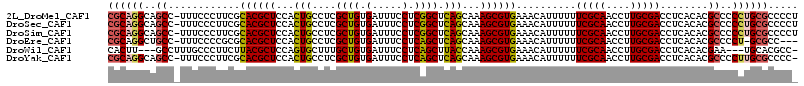
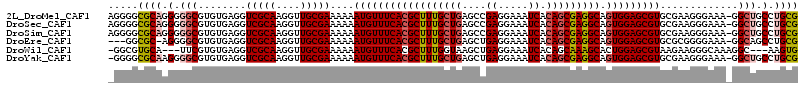
>2L_DroMel_CAF1 6186336 114 + 22407834 CGCAGGCAGCC-UUUCCCUUCGCACGCUCCACUGCCUCGCUGUGAUUUCCUCGGCUCAGCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGCCCCCUGCGCCCCU ((((((..((.-..........((((((....(((...((((.(.....).))))...))).))))))..........(((((....)))))........))..))))))..... ( -33.20) >DroSec_CAF1 22594 114 + 1 CGCAGGCAGCC-UUUCCCUUCGCACGCUCCACUGCCUCGCUGUGAUUUCCUCGGCUCAGCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGCCCCCUGCGCCCCU ((((((..((.-..........((((((....(((...((((.(.....).))))...))).))))))..........(((((....)))))........))..))))))..... ( -33.20) >DroSim_CAF1 32052 114 + 1 CGCAGGCAGCC-UUUCCCUUCGCACGCUCCACUGCCUCGCUGUGAUUUCCUCGGCUCAGCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGCCCCCUGCGCCCCU ((((((..((.-..........((((((....(((...((((.(.....).))))...))).))))))..........(((((....)))))........))..))))))..... ( -33.20) >DroEre_CAF1 22398 110 + 1 CGCAGGCUGCC-UUUCCCCGCGCACGCUCCACUGCCUCGCUGUGAUUUCCUCAGCUCAGCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGCCCCU-GCGCC--- ((((((..((.-..........((((((....(((...((((.(.....).))))...))).))))))..........(((((....)))))........)).)))-)))..--- ( -32.70) >DroWil_CAF1 890 108 + 1 CACUU---GCCUUUGCCCUUCUUACGCUCCAGUGCUUUGCUGUGAUUUCCUCAGCUUACCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGAA---UGCACGCC- .....---.......................((((((((.(((((.....(((((((....)))).))).........(((((....)))))..)))))))))---.))))...- ( -24.00) >DroYak_CAF1 33597 113 + 1 CGCAGGCAGCC-UUUCCCUUCGCACGCUCCACUGCCUCGCUGUGAUUUCCUCAGCUCAGCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGCCCCUUGCGCCCC- ((((((..((.-..........((((((....(((...((((.(.....).))))...))).))))))..........(((((....)))))........))..))))))....- ( -31.20) >consensus CGCAGGCAGCC_UUUCCCUUCGCACGCUCCACUGCCUCGCUGUGAUUUCCUCAGCUCAGCAAAGCGUGAAACAUUUUUUCGCAACCUUGCGACCUCACACGCCCCCUGCGCCCC_ ((((((..((............((((((...(((....((((.(.....).)))).)))...))))))..........(((((....)))))........))..))))))..... (-27.68 = -27.75 + 0.07)
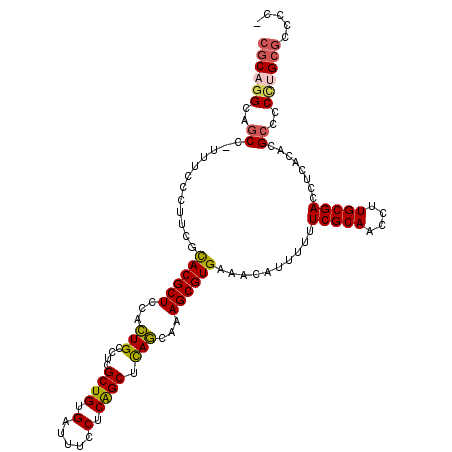
| Location | 6,186,336 – 6,186,450 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -32.74 |
| Energy contribution | -33.57 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
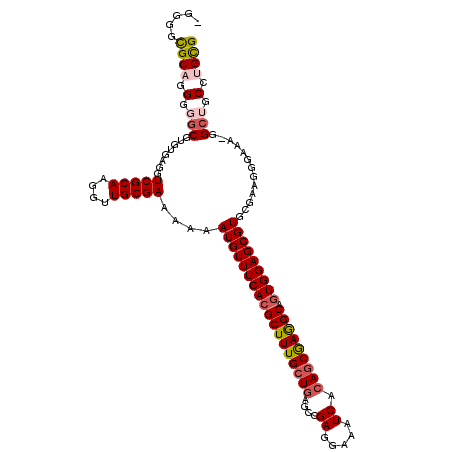
>2L_DroMel_CAF1 6186336 114 - 22407834 AGGGGCGCAGGGGGCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGCUGAGCCGAGGAAAUCACAGCGAGGCAGUGGAGCGUGCGAAGGGAAA-GGCUGCCUGCG .....((((((.(((........(((((....)))))....((((((((((((((((((....((.....)).))))))))).)))))))))...........-.))).)))))) ( -45.70) >DroSec_CAF1 22594 114 - 1 AGGGGCGCAGGGGGCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGCUGAGCCGAGGAAAUCACAGCGAGGCAGUGGAGCGUGCGAAGGGAAA-GGCUGCCUGCG .....((((((.(((........(((((....)))))....((((((((((((((((((....((.....)).))))))))).)))))))))...........-.))).)))))) ( -45.70) >DroSim_CAF1 32052 114 - 1 AGGGGCGCAGGGGGCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGCUGAGCCGAGGAAAUCACAGCGAGGCAGUGGAGCGUGCGAAGGGAAA-GGCUGCCUGCG .....((((((.(((........(((((....)))))....((((((((((((((((((....((.....)).))))))))).)))))))))...........-.))).)))))) ( -45.70) >DroEre_CAF1 22398 110 - 1 ---GGCGC-AGGGGCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGCUGAGCUGAGGAAAUCACAGCGAGGCAGUGGAGCGUGCGCGGGGAAA-GGCAGCCUGCG ---..(((-(((.((.((((....))))...(((((.....((((((((((((((((((...(((.....)))))))))))).))))))))).))))).....-.))..)))))) ( -46.20) >DroWil_CAF1 890 108 - 1 -GGCGUGCA---UUCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGGUAAGCUGAGGAAAUCACAGCAAAGCACUGGAGCGUAAGAAGGGCAAAGGC---AAGUG -.((.(((.---(((..(((((((((((....))))).......))))))((((..((..((((..(....).)))).....))..))))....)))..)))...))---..... ( -29.71) >DroYak_CAF1 33597 113 - 1 -GGGGCGCAAGGGGCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGCUGAGCUGAGGAAAUCACAGCGAGGCAGUGGAGCGUGCGAAGGGAAA-GGCUGCCUGCG -.((((((..(..((((......(((((....)))))....))))..)((((((..(((.((((..(....).))))....)))..))))))...........-.)).))))... ( -43.40) >consensus _GGGGCGCAGGGGGCGUGUGAGGUCGCAAGGUUGCGAAAAAAUGUUUCACGCUUUGCUGAGCCGAGGAAAUCACAGCGAGGCAGUGGAGCGUGCGAAGGGAAA_GGCUGCCUGCG .....((((.(.(((........(((((....)))))....((((((((((((((((((....((.....)).))))))))).))))))))).............))).).)))) (-32.74 = -33.57 + 0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:16 2006