
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
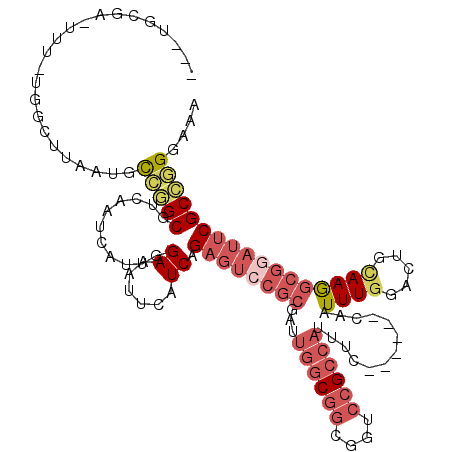
| Location | 6,178,516 – 6,178,625 |
| Length | 109 |
| Max. P | 0.746037 |

| Location | 6,178,516 – 6,178,625 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.96 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -21.46 |
| Energy contribution | -23.30 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6178516 109 + 22407834 ---UGCGA-UUU-CGGCUUAAUGCCGGCGUCAAUCAUUGGAAAUUCAUCAGAGGCCGCGAUUGGCGGCGGUCCGCCAUUUC------CAAUUUGGACUGCAAGGCGGAUUCGCCGGAAAA ---.....-(((-((((......(((.((((((((..(((...(((....))).))).)))))))).)))((((((...((------(.....)))......))))))...))))))).. ( -43.60) >DroPse_CAF1 36743 105 + 1 G--UGUGAAUAUCUUGAUUAAUGCGAGCGGCAAUUAUUGGAAAUUCAUCAGCGUCGUC------CGCAGGGCAGACAACUCAACCAAAAAUUUGGAC---AAAAUGUAUUCGCUUG---- .--.((((((((.(((.....((((..((((......(((....))).....))))..------))))(((.......)))..((((....)))).)---))...))))))))...---- ( -23.10) >DroSec_CAF1 14842 109 + 1 ---UGCGA-UUA-UGGUUUAAUGCCGGCGUCAAUCAUUGGAAAUUCAUCAGAGUCCGCGAUUGGCGGCGGUCCGCCAUUUC------CAAUUUGGACUGCAAGGCGGAUUCGCCGGGAAG ---.((((-...-..........(((.((((((((..((((...((....)).)))).)))))))).)))((((((...((------(.....)))......)))))).))))....... ( -38.00) >DroSim_CAF1 24313 98 + 1 ---UGCGA-UU------------UCAGCUUCAAUCAUUGGAAAUUCAUCAGAGUCCGCGAUUGGCGGCGGUCCGCCAUUUC------CAAUUUGGACUGCAAGGCGGAUUCGCCGGGAAG ---.....-((------------((....((((((..((((...((....)).)))).))))))((((((((((((...((------(.....)))......)))))).)))))))))). ( -33.20) >DroEre_CAF1 14612 109 + 1 ---UGCGA-UUUCUGGCUCAAUGCCGGCGUCAAUUAUUCGAAAUUCAUCGGAGU-CGCGAUUGGCGGCGGUCCGCCAUUUC------CAAUUUGGACUGUAAGGCGGAUUCGCCGGGAAA ---.....-((((((((......(((.((((((((.(((((......)))))..-...)))))))).)))((((((...((------(.....)))......))))))...)))))))). ( -42.40) >DroYak_CAF1 25639 113 + 1 AUGUGCCA-UUUCUAGCUCAAUGCAGGCGUCAAUUAUUCGAAAUUAUUCAGACUCCGCGAUUGGCGGCAGUCCGCCAUUUC------CAAUUUGGACUGCAAGGCGGAUUCGCCGGGAAA ........-(((((.((.((((((.((.(((...................))).)))).))))))(((((((((((...((------(.....)))......)))))))).)))))))). ( -34.81) >consensus ___UGCGA_UUU_UGGCUUAAUGCCGGCGUCAAUCAUUGGAAAUUCAUCAGAGUCCGCGAUUGGCGGCGGUCCGCCAUUUC______CAAUUUGGACUGCAAGGCGGAUUCGCCGGGAAA .......................(((((...........((......)).((((((((...((((((....)))))).............((((.....))))))))))))))))).... (-21.46 = -23.30 + 1.84)

| Location | 6,178,516 – 6,178,625 |
|---|---|
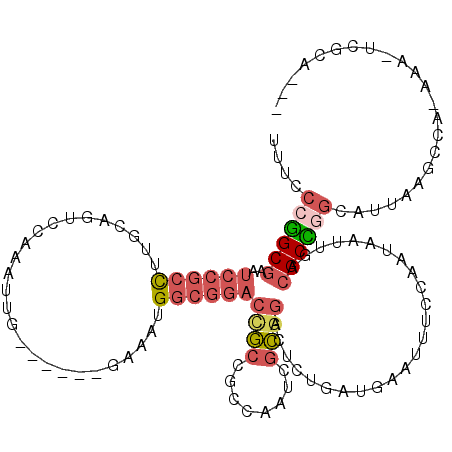
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.96 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -16.99 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6178516 109 - 22407834 UUUUCCGGCGAAUCCGCCUUGCAGUCCAAAUUG------GAAAUGGCGGACCGCCGCCAAUCGCGGCCUCUGAUGAAUUUCCAAUGAUUGACGCCGGCAUUAAGCCG-AAA-UCGCA--- ......((((....)))).(((.(((...((((------(((((..((((..(((((.....))))).))))....)))))))))....)))..((((.....))))-...-..)))--- ( -41.30) >DroPse_CAF1 36743 105 - 1 ----CAAGCGAAUACAUUUU---GUCCAAAUUUUUGGUUGAGUUGUCUGCCCUGCG------GACGACGCUGAUGAAUUUCCAAUAAUUGCCGCUCGCAUUAAUCAAGAUAUUCACA--C ----..(((...........---..((((....))))....(((((((((...)))------)))))))))..(((((.((.......(((.....)))........)).)))))..--. ( -22.76) >DroSec_CAF1 14842 109 - 1 CUUCCCGGCGAAUCCGCCUUGCAGUCCAAAUUG------GAAAUGGCGGACCGCCGCCAAUCGCGGACUCUGAUGAAUUUCCAAUGAUUGACGCCGGCAUUAAACCA-UAA-UCGCA--- ....(((((......((...)).(((...((((------(((((..((((...((((.....))))..))))....)))))))))....))))))))..........-...-.....--- ( -34.00) >DroSim_CAF1 24313 98 - 1 CUUCCCGGCGAAUCCGCCUUGCAGUCCAAAUUG------GAAAUGGCGGACCGCCGCCAAUCGCGGACUCUGAUGAAUUUCCAAUGAUUGAAGCUGA------------AA-UCGCA--- .......((((.((.((....(((((...((((------(((((..((((...((((.....))))..))))....))))))))))))))..)).))------------..-)))).--- ( -31.90) >DroEre_CAF1 14612 109 - 1 UUUCCCGGCGAAUCCGCCUUACAGUCCAAAUUG------GAAAUGGCGGACCGCCGCCAAUCGCG-ACUCCGAUGAAUUUCGAAUAAUUGACGCCGGCAUUGAGCCAGAAA-UCGCA--- .....(((((..((((((......(((.....)------))...)))))).)))))......(((-(...(((......))).............(((.....))).....-)))).--- ( -35.40) >DroYak_CAF1 25639 113 - 1 UUUCCCGGCGAAUCCGCCUUGCAGUCCAAAUUG------GAAAUGGCGGACUGCCGCCAAUCGCGGAGUCUGAAUAAUUUCGAAUAAUUGACGCCUGCAUUGAGCUAGAAA-UGGCACAU ......((((....)))).(((((..((((((.------(((((..((((((.((((.....))))))))))....))))).)))..)))....))))).((.((((....-)))).)). ( -38.70) >consensus UUUCCCGGCGAAUCCGCCUUGCAGUCCAAAUUG______GAAAUGGCGGACCGCCGCCAAUCGCGGACUCUGAUGAAUUUCCAAUAAUUGACGCCGGCAUUAAGCCA_AAA_UCGCA___ ....((((((..((((((..........................))))))((((........)))).........................))))))....................... (-16.99 = -18.10 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:06 2006