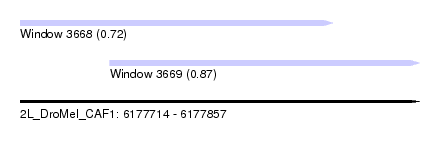
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,177,714 – 6,177,857 |
| Length | 143 |
| Max. P | 0.868736 |

| Location | 6,177,714 – 6,177,826 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -29.88 |
| Energy contribution | -34.32 |
| Covariance contribution | 4.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6177714 112 + 22407834 --------CCGCUGAUUUCUUUGACUGCCGGGCAGGCGAACAAGCCGUUUGACAGACAGCGCAUGCCCAACGGCUUUGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGGCAAUGCUU --------..((..((((((....(((.(((((.(((......)))))))).)))..(((.((((((((.((((....))))...)))))))).)))......))))))..))....... ( -46.00) >DroSec_CAF1 14096 104 + 1 --------CCGCUGAUUUGUUUGACUGCCGGGCAGGCGAACAAGCCGUUUGACAGACAGCGCAUGCCCAACGGCUUCGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGG-------- --------((((((.((((((.((((((...)))(((......)))))).)))))))))).((((((((.((((....))))...))))))))..........)).......-------- ( -44.00) >DroSim_CAF1 23572 104 + 1 --------CCGCUGAUUUGUUUGAUGGCCGGGCAGGCGAACAAGCCGUUUGAUAGACAGCGCAUGCCCAACGGCUUCGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGG-------- --------((((((.((((((.((((((..(.........)..)))))).)))))))))).((((((((.((((....))))...))))))))..........)).......-------- ( -42.70) >DroEre_CAF1 13814 101 + 1 --------CCGCUGAUUUGUUUAACUGCC--------UAACAUGCCGCUUGACAGACAGCGCAUGCCCAACGGCUUCGG--GUACUGGGUAUGAGUUAUGGUUGGAAGUUGGCAAUGCC- --------..((..((((.(......(((--------((((....((((........))))((((((((..(.(....)--.)..)))))))).)))).))).).))))..))......- ( -30.70) >DroYak_CAF1 24704 119 + 1 CGGCUCCUCCGCUGAUUUGUUUAACUGCCGGGCAGGCAAACAAGCUGCUUGACAGACAGCGCAUGCCCAACUGCUUCGGCUGCACUGGGUAUGAGUUAUGGCUGGAAGUUGGCAAUGCC- .(((......((..((((......(((.(((((((.(......)))))))).))).((((.((((((((...((....)).....)))))))).......)))).))))..))...)))- ( -42.20) >consensus ________CCGCUGAUUUGUUUGACUGCCGGGCAGGCGAACAAGCCGUUUGACAGACAGCGCAUGCCCAACGGCUUCGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGGCAAUGC__ ..........((..((((......(((.(((((.(((......)))))))).)))..(((.((((((((.((((....))))...)))))))).)))........))))..))....... (-29.88 = -34.32 + 4.44)

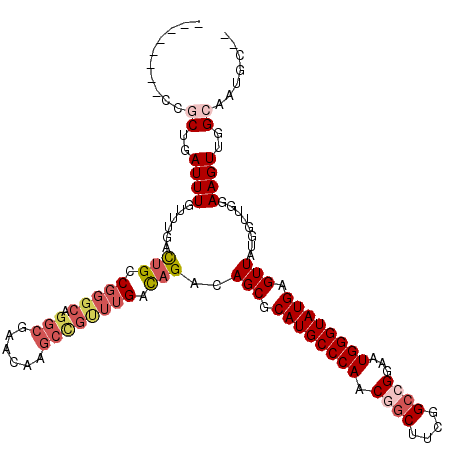
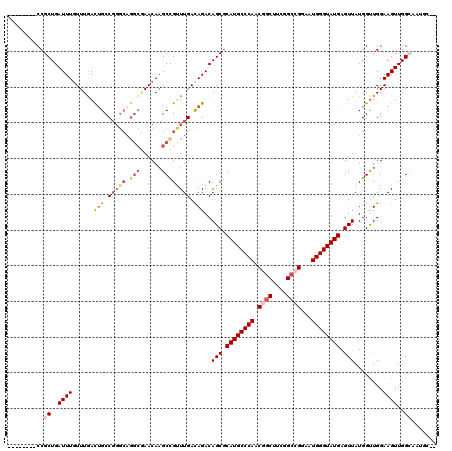
| Location | 6,177,746 – 6,177,857 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.36 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -27.52 |
| Energy contribution | -30.78 |
| Covariance contribution | 3.26 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6177746 111 + 22407834 CAAGCCGUUUGACAGACAGCGCAUGCCCAACGGCUUUGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGGCAAUGCUUCUGCUGUCUGCUGUUUGUUUGUACACUCGG----A (((((.....(.((((((((.((((((((.((((....))))...))))))))..........((((((.......))))))))))))))).....))))).........----. ( -43.00) >DroSec_CAF1 14128 100 + 1 CAAGCCGUUUGACAGACAGCGCAUGCCCAACGGCUUCGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGG-----------CUGUCUGCUGUUGGUUUGCACACUCGG----A (((((((...(.((((((((.((((((((.((((....))))...))))))))..(((....)))......)-----------))))))))...))))))).........----. ( -43.20) >DroSim_CAF1 23604 100 + 1 CAAGCCGUUUGAUAGACAGCGCAUGCCCAACGGCUUCGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGG-----------CUGUCUGCUGUUUGUUUGCACACUCGG----A ....(((...((((((((((.((((((((.((((....))))...))))))))....(((((..(...)..)-----------))))..))))))))))........)))----. ( -40.00) >DroEre_CAF1 13838 106 + 1 CAUGCCGCUUGACAGACAGCGCAUGCCCAACGGCUUCGG--GUACUGGGUAUGAGUUAUGGUUGGAAGUUGGCAAUGCC---GCUGUCUGCCGGUUGUCUGCGCACUCCG----A ......((..(.((((((((.((((((((..(.(....)--.)..)))))))).....((((.(..(((.(((...)))---)))..).)))))))))))))))......----. ( -37.40) >DroYak_CAF1 24744 112 + 1 CAAGCUGCUUGACAGACAGCGCAUGCCCAACUGCUUCGGCUGCACUGGGUAUGAGUUAUGGCUGGAAGUUGGCAAUGCC---ACUGUUGGCCGUUUGUUUGCACACUUUGCCAUU ...(.(((..((((((((((.((((((((...((....)).....)))))))).)))..(((..(.(((.(((...)))---))).)..)))))))))).))))........... ( -43.10) >consensus CAAGCCGUUUGACAGACAGCGCAUGCCCAACGGCUUCGGCCGGAAUGGGUAUGAGUUAUGGUUGGAAGUUGGCAAUGC_____CUGUCUGCUGUUUGUUUGCACACUCGG____A ......((..((((((((((.((((((((.((((....))))...)))))))).)))..(((.((.(((.(((...)))...))).)).)))))))))).))............. (-27.52 = -30.78 + 3.26)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:03 2006