
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,175,224 – 6,175,368 |
| Length | 144 |
| Max. P | 0.997960 |

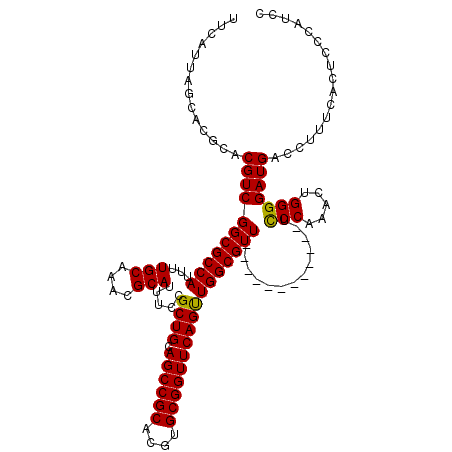
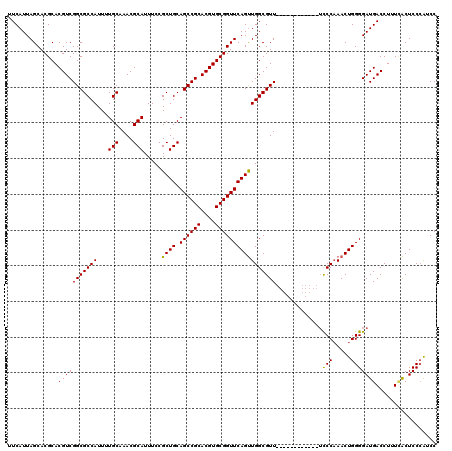
| Location | 6,175,224 – 6,175,332 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -33.14 |
| Energy contribution | -32.82 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6175224 108 + 22407834 UUCAUUAGCACGCACGUCGGCGCCAUUUUGCAAACGCAUUUCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU------------UCCCAAACUGGGGAUGACCUUUUACUCCCAUCC ......(((.(((((((((((((......))....(((.......))).)))).))))))))))(((((((....------------..)).)))))(((((((....))).)))).... ( -35.30) >DroSec_CAF1 11538 108 + 1 UUCAUUAGCACGCACGUCGGCGCCAUUUUGCAAACGCAUUUCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU------------UCCCAAACUGGGGAUGACCUUUCACUCCCAUCC ......(((.(((((((((((((......))....(((.......))).)))).))))))))))(((((((....------------..)).)))))(((((((....))).)))).... ( -37.50) >DroSim_CAF1 20965 108 + 1 UUCAUUAGCACGCACGUCGGCGCCAUUUUGCAAACGCAUUUCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU------------UCCCAAACUGGGGAUGACCUUUCACUCCCAUCC ......(((.(((((((((((((......))....(((.......))).)))).))))))))))(((((((....------------..)).)))))(((((((....))).)))).... ( -37.50) >DroEre_CAF1 11012 107 + 1 UUCAUUAGCACGCACGUCGGCGCCAUUUUGCAAACGCAUUUCCGCUGCAGCCGCACGUGCGGUUCAGCUGGCGUU-------------UCCAAACUGGGGAUGACCUUUGGCUCCCGGCC ...............(((((.((((...(((....))).....((((.((((((....)))))))))).(((((.-------------.((.....))..))).))..))))..))))). ( -40.70) >DroYak_CAF1 21857 120 + 1 UUCAUUAGCACGCACGUCGGCGCCAUUUUGCAAACGCAUUUCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUUCAACAUCCAACAUCCCCAACUGGGGAUGACCUUUCACUCCAAGCC .......((..((((((((((((......))....(((.......))).)))).))))))(((...((((.....))))......(((((((....))))))))))...........)). ( -42.10) >consensus UUCAUUAGCACGCACGUCGGCGCCAUUUUGCAAACGCAUUUCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU____________UCCCAAACUGGGGAUGACCUUUCACUCCCAUCC ..............(((((((((((...(((....))).....((((.((((((....))))))))))))))))).............(((.....)))))))................. (-33.14 = -32.82 + -0.32)

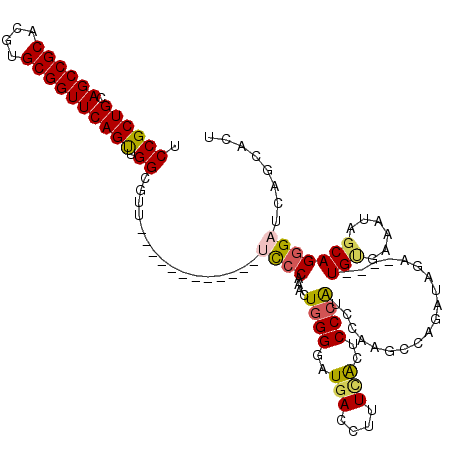
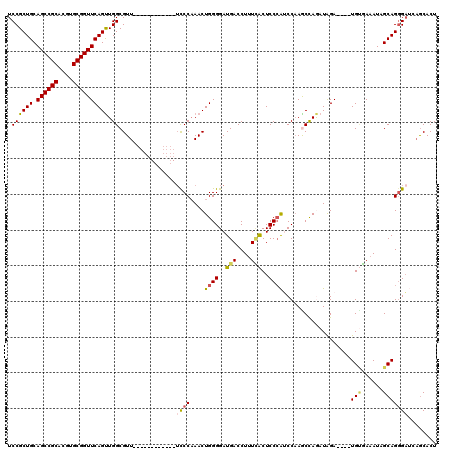
| Location | 6,175,264 – 6,175,368 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6175264 104 + 22407834 UCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU------------UCCCAAACUGGGGAUGACCUUUUACUCCCAUCCAAGCCAGAUAGA----UGUGAAAUAGCAGUGAUCAGCACU ..((((((((((((....))))))...(((((...------------..((.....))(((((............)))))..))))).....----.........))))))......... ( -32.20) >DroSec_CAF1 11578 104 + 1 UCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU------------UCCCAAACUGGGGAUGACCUUUCACUCCCAUCCAAACCAGAUAGA----UGUGAAAUAGCAGGGAUCAGCACU .(.((((.((((((....)))))))))).)((...------------((((...(((((((((............)))))...)))).....----(((......)))))))...))... ( -34.70) >DroSim_CAF1 21005 104 + 1 UCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU------------UCCCAAACUGGGGAUGACCUUUCACUCCCAUCCAAACCAGAUAGA----UGUGAAAUAGCAGGGAUCAGCACU .(.((((.((((((....)))))))))).)((...------------((((...(((((((((............)))))...)))).....----(((......)))))))...))... ( -34.70) >DroEre_CAF1 11052 107 + 1 UCCGCUGCAGCCGCACGUGCGGUUCAGCUGGCGUU-------------UCCAAACUGGGGAUGACCUUUGGCUCCCGGCCAAGCCAAAUGGAUCCAUGGCACAUCGCAGGGAAGCGCACU ...((((.((((((....))))))))))..(((((-------------(((......(.((((....(((((.....)))))((((..((....)))))).)))).)..))))))))... ( -48.20) >DroYak_CAF1 21897 120 + 1 UCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUUCAACAUCCAACAUCCCCAACUGGGGAUGACCUUUCACUCCAAGCCCAGCUAAAUAGAUCCAUGCGAAAUCGCAGGGCAAAACACU .((((((.((((((....))))))))).))).(((..........(((((((....)))))))..............((((..((....)).....((((....))))))))..)))... ( -41.30) >consensus UCCGCUGCAGCCGCACGUGCGGUUCAGUUGGCGUU____________UCCCAAACUGGGGAUGACCUUUCACUCCCAUCCAAGCCAGAUAGA____UGUGAAAUAGCAGGGAUCAGCACU .((((((.((((((....)))))))))).))................((((....((((..(((....)))..))))...................(((......)))))))........ (-25.10 = -25.42 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:01 2006