
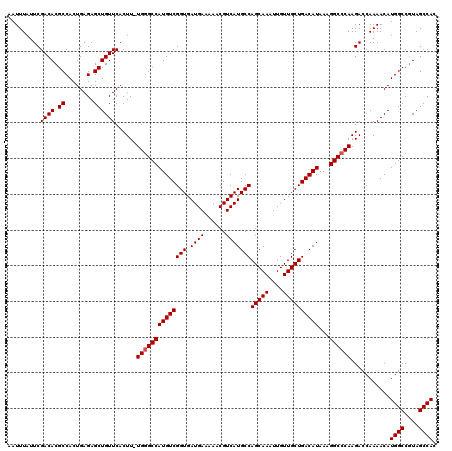
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,173,348 – 6,173,506 |
| Length | 158 |
| Max. P | 0.999627 |

| Location | 6,173,348 – 6,173,467 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.99 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -31.72 |
| Energy contribution | -31.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6173348 119 + 22407834 AAUUUAUUCGACACGCCACUGAGAGCUGUUCACUU-UGUGCCAUGUCGGUGAUGAAAAACGUCAUGCCAGCAAAUUGUUGCUGACAUAAAGGCCCAAGACCAAAACAUGGCCGUAGCCAC .........((((.(.(((.(((.........)))-.))).).))))(((((((.....))))(((.((((((....)))))).)))...((((..............))))...))).. ( -28.64) >DroSec_CAF1 9676 119 + 1 AAUUUAUUCGACACGCCACUGAGAGCUGUUCACUU-UGGGCCAUGUCGGUGAUGAAAAACGUCAUGCCAGCAAAUUGUUGCUGACAUAAAGGCCCAAGACCAAAACAUGGCCGUAGCCAC .........((((.((........))))))...((-((((((((((((((.((((......)))))))(((((....))))))))))...)))))))).........((((....)))). ( -34.80) >DroSim_CAF1 19052 119 + 1 AAUUUAUUCGACACGCCACUGAGAGCUGUUCACUU-UGGGCCAUGUCGGUGAUGAAAAACGUCAUGCCAGCAAAUUGUUGCUGACAUAAAGGCCCAAGACCAAAACAUGGCCGUAGCCAC .........((((.((........))))))...((-((((((((((((((.((((......)))))))(((((....))))))))))...)))))))).........((((....)))). ( -34.80) >DroEre_CAF1 9144 119 + 1 AAUUUAUUCGACACGCCACUGAGAGCUGUUCACUU-UGGGCCAUGUCGGUGAUGAAAAACGUCAUGCCAGCAAAUUGUUGCUGACAUAAAGGCCCAAGACCAAAACAUGGCCGUAGCCAC .........((((.((........))))))...((-((((((((((((((.((((......)))))))(((((....))))))))))...)))))))).........((((....)))). ( -34.80) >DroYak_CAF1 19367 120 + 1 AAUUUAUUCGACACGCCACUGAGAGCUGUUCACUUUUGGGCCAUGUCGGUGAUGAAAAACGUCAUGCCAGCAAAUUGUUGCUGACAUAAAGGCCCAAGACCAAAACAUGGCCAUAGCCAC .........((((.((........))))))...(((((((((((((((((.((((......)))))))(((((....))))))))))...)))))))))........((((....)))). ( -35.80) >consensus AAUUUAUUCGACACGCCACUGAGAGCUGUUCACUU_UGGGCCAUGUCGGUGAUGAAAAACGUCAUGCCAGCAAAUUGUUGCUGACAUAAAGGCCCAAGACCAAAACAUGGCCGUAGCCAC .........((((.((........))))))......((((((((((((((.((((......)))))))(((((....))))))))))...))))))...........((((....)))). (-31.72 = -31.92 + 0.20)


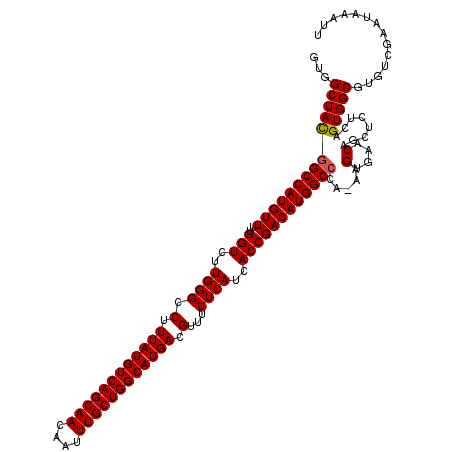
| Location | 6,173,348 – 6,173,467 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.99 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -40.58 |
| Energy contribution | -40.62 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6173348 119 - 22407834 GUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGGCACA-AAGUGAACAGCUCUCAGUGGCGUGUCGAAUAAAUU ..((((..(((((.....)))))...)))).((((((((((((....)))))))))))).............(((((((.(((.-.((.(.....).))..))).)))))))........ ( -44.10) >DroSec_CAF1 9676 119 - 1 GUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGGCCCA-AAGUGAACAGCUCUCAGUGGCGUGUCGAAUAAAUU ...((((((((((((((..(((..((((.(.((((((((((((....)))))))))))).)...))))..))))))))))))..-..(....)........))))).............. ( -42.10) >DroSim_CAF1 19052 119 - 1 GUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGGCCCA-AAGUGAACAGCUCUCAGUGGCGUGUCGAAUAAAUU ...((((((((((((((..(((..((((.(.((((((((((((....)))))))))))).)...))))..))))))))))))..-..(....)........))))).............. ( -42.10) >DroEre_CAF1 9144 119 - 1 GUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGGCCCA-AAGUGAACAGCUCUCAGUGGCGUGUCGAAUAAAUU ...((((((((((((((..(((..((((.(.((((((((((((....)))))))))))).)...))))..))))))))))))..-..(....)........))))).............. ( -42.10) >DroYak_CAF1 19367 120 - 1 GUGGCUAUGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGGCCCAAAAGUGAACAGCUCUCAGUGGCGUGUCGAAUAAAUU ..((((..(((((.....)))))...)))).((((((((((((....)))))))))))).............(((((((.(((...(((.....)))....).)))))))))........ ( -40.70) >consensus GUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGGCCCA_AAGUGAACAGCUCUCAGUGGCGUGUCGAAUAAAUU ...((((((((((((((..(((..((((.(.((((((((((((....)))))))))))).)...))))..)))))))))))).....(....)........))))).............. (-40.58 = -40.62 + 0.04)

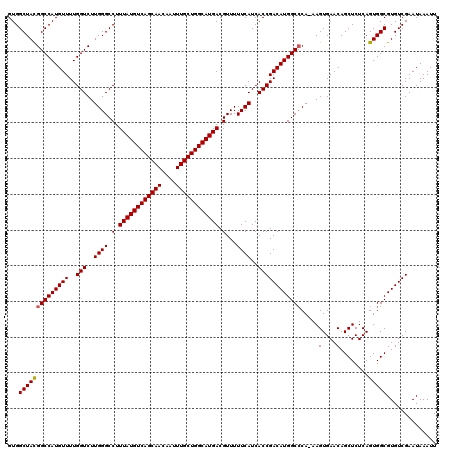
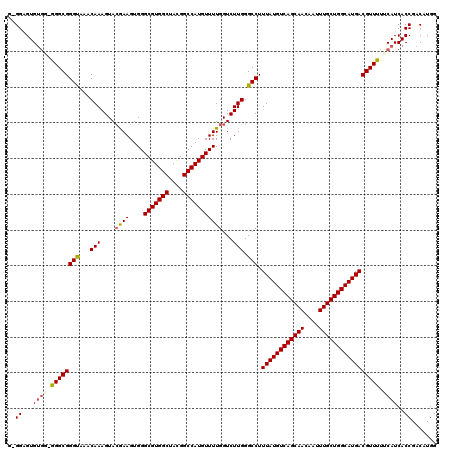
| Location | 6,173,387 – 6,173,506 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.04 |
| Mean single sequence MFE | -45.26 |
| Consensus MFE | -39.32 |
| Energy contribution | -39.60 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6173387 119 - 22407834 GCGGGGUGUGG-GGGCGGGUAAACAAAGUACGAAGUGGGCGUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGG .(((((((.((-((((((((...(((.(.((.(((...(((((((....)))))))))).)))))).))).((((((((((((....))))))))))))))))))))))).)))...... ( -47.20) >DroSec_CAF1 9715 118 - 1 --GGAGUGUGUGGGGCGGGUAAACAAAGUAGGAAGUGGGCGUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGG --.....((((.((...((.((((....(((((...(((((((((....)))))))))...))))).....((((((((((((....)))))))))))).)))).))....)).)))).. ( -44.80) >DroSim_CAF1 19091 117 - 1 --GGAGUGUAA-GGGCGGGUAAACAAAGUACGAAGUGGGCGUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGG --((.(((.((-((((((((...(((.(.((.(((...(((((((....)))))))))).)))))).))).((((((((((((....))))))))))))))))))))))..))....... ( -42.70) >DroEre_CAF1 9183 119 - 1 GGGGAGUGUGG-AGGCGGGUAUACAAAGUACAAAGUGGGCGUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGG .((..(((.((-((((((((...(((.(.((.....(((((((((....)))))))))..)))))).))).((((((((((((....))))))))))))))))))))))..))....... ( -45.50) >DroYak_CAF1 19407 118 - 1 GUGGGGUG-GU-AGGCGGGCAUACAAAGUACAAAGUGGGCGUGGCUAUGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGG ((((((((-(.-((((((((...(((.(.((.....(((((((((....)))))))))..)))))).))).((((((((((((....))))))))))))))))).))))).)).)).... ( -46.10) >consensus G_GGAGUGUGG_GGGCGGGUAAACAAAGUACGAAGUGGGCGUGGCUACGGCCAUGUUUUGGUCUUGGGCCUUUAUGUCAGCAACAAUUUGCUGGCAUGACGUUUUUCAUCACCGACAUGG ..((...((((.((((((((...(((....((((....(((((((....)))))))))))...))).))).((((((((((((....))))))))))))))))).))))..))....... (-39.32 = -39.60 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:58 2006