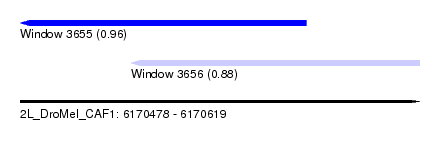
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,170,478 – 6,170,619 |
| Length | 141 |
| Max. P | 0.958866 |

| Location | 6,170,478 – 6,170,579 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6170478 101 - 22407834 GCAUUGUGUGUGCACACACACA----CAUACACUCUCUUACCGACAACGACAGCUUGUUGUCACAUUUACAAUAUCCUUCAGAGUUUAUUCAAGCACACAGAGAG ....((((((((....))))))----))....((((((....((((((((....)))))))).....................((((....))))....)))))) ( -27.40) >DroSec_CAF1 6758 103 - 1 GCAUUGUGUGUGCACA--CACACUACGAUAUACACACUUACCGACAACGACAGCUUGUUGUCAAAUUUACAAUAUCCUUCAGAGUUUAUUCAAGCACACAGAGAG ...((.(((((((...--........................((((((((....))))))))........((((..((....))..))))...))))))).)).. ( -22.50) >DroSim_CAF1 16153 105 - 1 GCAUUGUGUGUGCACACACACACUGCCAUAUACACACUUACCGACAACGACAGCUUGUUGUCACAUUUACAAUAUCCUUCAGAGUUUAUUCAAGCACACAGAGAG (((.(((((((....))))))).)))..........(((...((((((((....)))))))).....................((((....)))).....))).. ( -23.70) >consensus GCAUUGUGUGUGCACACACACACU_CCAUAUACACACUUACCGACAACGACAGCUUGUUGUCACAUUUACAAUAUCCUUCAGAGUUUAUUCAAGCACACAGAGAG ...((.(((((((.............................((((((((....))))))))........((((..((....))..))))...))))))).)).. (-22.50 = -22.50 + 0.00)

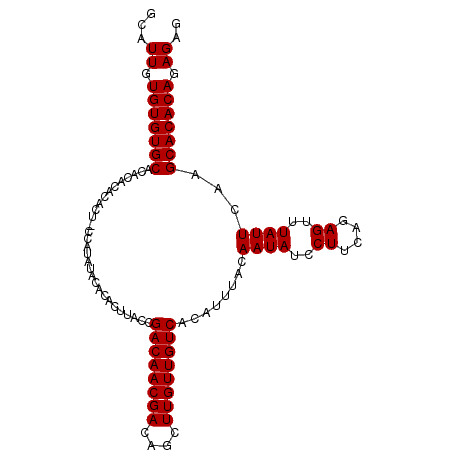
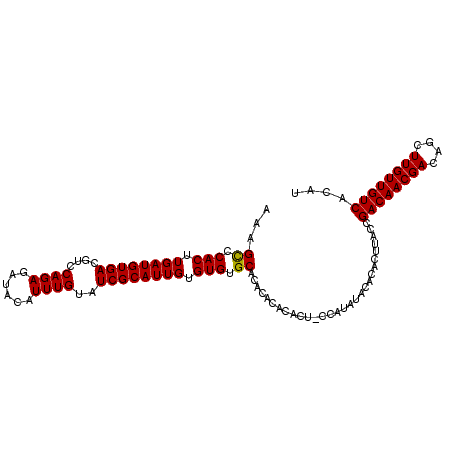
| Location | 6,170,517 – 6,170,619 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.14 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6170517 102 - 22407834 AAAGUCCACUUGAUGUGACGUCCAGAGAUACAUUUGUAUCGCAUUGUGUGUGCACACACACA----CAUACACUCUCUUACCGACAACGACAGCUUGUUGUCACAU ............((((((((((..(.(((((....))))).)..((((((((....))))))----))..............)))(((((....)))))))))))) ( -29.10) >DroSec_CAF1 6797 104 - 1 AAAGCCCACUUGAUGUGACGUCCAGAGAUACAUUUGUAUCGCAUUGUGUGUGCACA--CACACUACGAUAUACACACUUACCGACAACGACAGCUUGUUGUCAAAU ...((.(((.....))).........(((((....)))))))((((((((((....--))))).))))).............((((((((....)))))))).... ( -27.00) >DroSim_CAF1 16192 106 - 1 AAAGCCCACUUGAUGUGACGUCCAGAGAUACAUUUGUAUCGCAUUGUGUGUGCACACACACACUGCCAUAUACACACUUACCGACAACGACAGCUUGUUGUCACAU ............((((((((((....(((((....)))))(((.(((((((....))))))).)))................)))(((((....)))))))))))) ( -31.60) >consensus AAAGCCCACUUGAUGUGACGUCCAGAGAUACAUUUGUAUCGCAUUGUGUGUGCACACACACACU_CCAUAUACACACUUACCGACAACGACAGCUUGUUGUCACAU ...((.(((.((((((((....((((......))))..)))))))).))).)).............................((((((((....)))))))).... (-23.32 = -23.10 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:49 2006