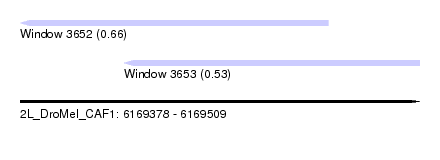
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,169,378 – 6,169,509 |
| Length | 131 |
| Max. P | 0.660698 |

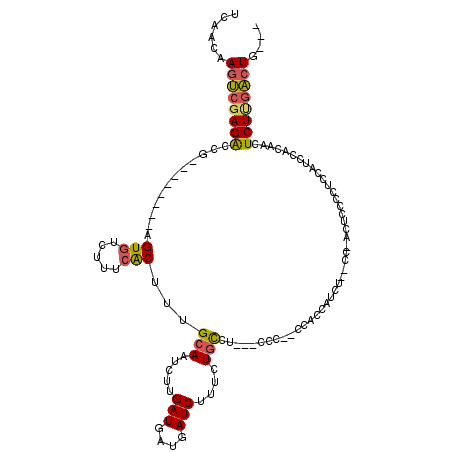
| Location | 6,169,378 – 6,169,479 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.91 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6169378 101 - 22407834 UCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUUUCUGCCA---UCC--CCACCAUCU--GC-ACUCCCCACCAUCUACAACUCUUGACUG--- ......((((((((..(--------((((.....)))))((((((..((..((((..........))---)).--.))..)).)--))-)..................)))))))).--- ( -18.50) >DroPse_CAF1 21899 115 - 1 UCAACGACGAGAGGCCGGAGUCGAGUGUGUCUUUGGCUUUGUAAUCAUGAUGAUGAUCUCUCUCUCU---CCCUGUCUCUGUCUCUCC-GUGCCAUCCCAUCCCCCACUCU-GUCUGGGG .....(((.((((((.((((..(((...((.....))...(..(((((....)))))..).))).))---))..))))))))).....-.............(((((....-...))))) ( -28.20) >DroSec_CAF1 5665 99 - 1 UCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAGUCUUGAUGAUGAUCUUUCUGCC---------UCACCAUCCCCCC-ACUCCCCUCCAUCCACAACUCUUGACUG--- ......((((((((..(--------((((.....))))).((((....(((....)))...)))).---------.............-...................)))))))).--- ( -19.10) >DroSim_CAF1 15050 100 - 1 UCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUUUCUGCC---------UCACUAUCCCCCCUACUCCCCUCCAUCCACAACUCUUGACUG--- ......((((((((..(--------((((.....))))).(((.....(((....)))....))).---------.................................)))))))).--- ( -15.40) >DroEre_CAF1 4919 101 - 1 UCAACAAGUCGAGACCG--------AGUGUCUUUCACUUGGCAAUCUUGAUGAUUAUCUUUCUGCCU---CCC--CCUCCCCCU--CC-ACUCCCCGCCAUCCACCACUCUCGACUG--- ......(((((((((((--------((((.....))))))).((((.....))))............---...--.........--..-...................)))))))).--- ( -22.50) >DroYak_CAF1 15295 103 - 1 UCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUGCCUGCCUUUACCC--CCACUGCCU--CC-ACUCUUCUCCAUCCACCA-UCUUGACUG--- ......((((((((..(--------((((.....))))).((((((........))).)))............--.........--..-..................-)))))))).--- ( -16.90) >consensus UCAACAAGUCGAGACCG________AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUUUCUGCCU___CCC__CCACCAUCU__CC_ACUCCCCUCCAUCCACAACUCUUGACUG___ ......((((((((............(((.....)))...(((.....(((....)))....)))...........................................)))))))).... (-10.35 = -10.38 + 0.03)


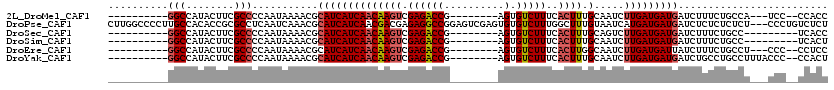
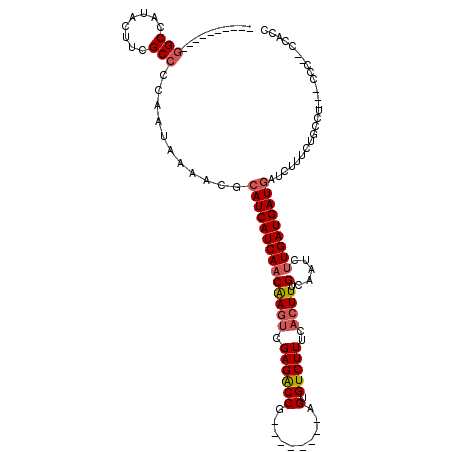
| Location | 6,169,412 – 6,169,509 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -13.58 |
| Energy contribution | -14.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6169412 97 - 22407834 ----------GGCCAUACUUCGCCCCAAUAAAACGCAUCAUCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUUUCUGCCA---UCC--CCACC ----------(((........((...........))(((((((.((((....(.(.(--------((((.....))))).))...)))).)))))))......))).---...--..... ( -19.50) >DroPse_CAF1 21937 117 - 1 CUUGGCCCCUUGCCACACCGCGCCUCAAUCAAACGCAUCAUCAACGACGAGAGGCCGGAGUCGAGUGUGUCUUUGGCUUUGUAAUCAUGAUGAUGAUCUCUCUCUCU---CCCUGUCUCU ..((((.....))))....................((((((((.......((((((((((.((....)).)))))))))).......))))))))............---.......... ( -28.44) >DroSec_CAF1 5701 93 - 1 ----------GGCCAUACUUCGCCCCAAUAAAACGCAUCAUCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAGUCUUGAUGAUGAUCUUUCUGCC---------UCACC ----------(((........((...........))(((((((.....(((((((.(--------((((.....)))))....))))))))))))))......)))---------..... ( -21.30) >DroSim_CAF1 15087 93 - 1 ----------GGCCAUACUUCGCCCCAAUAAAACGCAUCAUCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUUUCUGCC---------UCACU ----------(((........((...........))(((((((.((((....(.(.(--------((((.....))))).))...)))).)))))))......)))---------..... ( -19.00) >DroEre_CAF1 4953 97 - 1 ----------GGCCAUACUUCGCCCCAAUAAAACGCAUCAUCAACAAGUCGAGACCG--------AGUGUCUUUCACUUGGCAAUCUUGAUGAUUAUCUUUCUGCCU---CCC--CCUCC ----------(((........)))..........(((..........((((((((((--------((((.....)))))))...)))))))...........)))..---...--..... ( -20.90) >DroYak_CAF1 15328 100 - 1 ----------GGCCAUACUUCGCCCCAAUAAAACGCAUCAUCAACAAGUCGAGACCG--------AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUGCCUGCCUUUACCC--CCACU ----------(((........)))..........(((((((((.((((....(.(.(--------((((.....))))).))...)))).))))))..)))............--..... ( -19.80) >consensus __________GGCCAUACUUCGCCCCAAUAAAACGCAUCAUCAACAAGUCGAGACCG________AGUGUCUUUCACUUUGCAAUCUUGAUGAUGAUCUUUCUGCCU___CCC__CCACC ..........(((........)))...........((((((((((((((.((((((..........).)))))..)))).).....)))))))))......................... (-13.58 = -14.13 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:47 2006