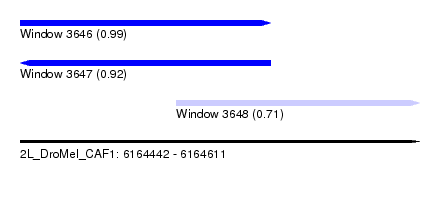
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,164,442 – 6,164,611 |
| Length | 169 |
| Max. P | 0.992607 |

| Location | 6,164,442 – 6,164,548 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -20.36 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6164442 106 + 22407834 UUGCCAAAAAUUCA-------AA----UC--GGAAGGCUUGUUAAGUUGCAAAAGUCGUGUCUAACAAUGGUAAGAAGG-UUUUUCAUCUGGGGUAGUGCGACUGCCCCUUUUUGUGGCA .(((((......((-------(.----.(--....)..)))...((.((.((((..(...(((.((....)).))).).-.)))))).))(((((((.....)))))))......))))) ( -29.90) >DroSec_CAF1 851 106 + 1 UUGCCACAUAUCCA-------AA----UA--GGAAGGCUUGUUGAGUUGCAAUAAUCGUGUCUAACAAUGGUAAGAAGG-UUUCCCAACUGGGGUAGUGCGACUGCCCCUUUUUGUGGCA .(((((((......-------..----..--(((((.(((.(((.((((................))))..))).))).-))))).....(((((((.....)))))))....))))))) ( -34.69) >DroSim_CAF1 10221 108 + 1 UUGCCACAAAUCCAA------AA----UA--GGAAGGCUUGUUGAGCUGCAAAAGUCGUGUCUAACAAUGGUAAGAAGGCUUUCCCAACUGGGGUAGUGCGACUGCCCCUUUUUGUGGCA .((((((((((((..------..----..--))).(((..((((.(((((.((((((...(((.((....)).))).))))))(((....)))))))).)))).)))....))))))))) ( -41.90) >DroEre_CAF1 311 95 + 1 UUGCCACAA----------AAAAACGUUAGUUGAGCAAAGCCUGAGUUGCAGAAGUUGU--------------AGGAGG-UUUCCCAACUGGGGUACUGCGACUGCCCCUUUUUGUGGCA .((((((((----------(((......(((((.(..((((((...((((((...))))--------------)).)))-))).))))))((((((.......))))))))))))))))) ( -33.30) >DroYak_CAF1 10314 119 + 1 UUGCCAUAAAACCUAAAAAAAAAACAUUAGUUGAAGGCUGGUUGAGUUGCAAAAGUUGUGUUUAACAAUGGUGAGAAGG-CUUCCCAUCUAGGGUAGUGCGACUGCCCCUUUUUGUGGCA .((((((((((..................((((((.((...(((.....))).....)).)))))).((((..((....-))..))))...((((((.....))))))..)))))))))) ( -33.10) >consensus UUGCCACAAAUCCA_______AA____UA__GGAAGGCUUGUUGAGUUGCAAAAGUCGUGUCUAACAAUGGUAAGAAGG_UUUCCCAACUGGGGUAGUGCGACUGCCCCUUUUUGUGGCA .(((((((((...................................................................((.....))....(((((((.....)))))))..))))))))) (-20.36 = -21.08 + 0.72)

| Location | 6,164,442 – 6,164,548 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.97 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6164442 106 - 22407834 UGCCACAAAAAGGGGCAGUCGCACUACCCCAGAUGAAAAA-CCUUCUUACCAUUGUUAGACACGACUUUUGCAACUUAACAAGCCUUCC--GA----UU-------UGAAUUUUUGGCAA (((((.(((..((((.((.....)).))))..........-...........(((((((................))))))).......--..----..-------....))).))))). ( -18.49) >DroSec_CAF1 851 106 - 1 UGCCACAAAAAGGGGCAGUCGCACUACCCCAGUUGGGAAA-CCUUCUUACCAUUGUUAGACACGAUUAUUGCAACUCAACAAGCCUUCC--UA----UU-------UGGAUAUGUGGCAA (((((((....((((.((.....)).))))((((((....-))........(((((.....)))))......))))..........(((--..----..-------.)))..))))))). ( -28.30) >DroSim_CAF1 10221 108 - 1 UGCCACAAAAAGGGGCAGUCGCACUACCCCAGUUGGGAAAGCCUUCUUACCAUUGUUAGACACGACUUUUGCAGCUCAACAAGCCUUCC--UA----UU------UUGGAUUUGUGGCAA (((((((((..((((.((.....)).))))(((((.((((((..(((.((....)).)))...).))))).)))))..........(((--..----..------..)))))))))))). ( -34.70) >DroEre_CAF1 311 95 - 1 UGCCACAAAAAGGGGCAGUCGCAGUACCCCAGUUGGGAAA-CCUCCU--------------ACAACUUCUGCAACUCAGGCUUUGCUCAACUAACGUUUUU----------UUGUGGCAA (((((((((((.((((((((((((......((..((....-))..))--------------.......))))......)))..)))))(((....))))))----------)))))))). ( -32.32) >DroYak_CAF1 10314 119 - 1 UGCCACAAAAAGGGGCAGUCGCACUACCCUAGAUGGGAAG-CCUUCUCACCAUUGUUAAACACAACUUUUGCAACUCAACCAGCCUUCAACUAAUGUUUUUUUUUUAGGUUUUAUGGCAA ((((((((((.((((.((.....)).)))).(((((..((-....))..)))))............)))))..........(((((..((..((....))..))..)))))...))))). ( -22.70) >consensus UGCCACAAAAAGGGGCAGUCGCACUACCCCAGUUGGGAAA_CCUUCUUACCAUUGUUAGACACGACUUUUGCAACUCAACAAGCCUUCC__UA____UU_______UGGAUUUGUGGCAA (((((((((..((((.((.....)).))))(((((.((((..........................)))).)))))..................................))))))))). (-18.73 = -19.97 + 1.24)

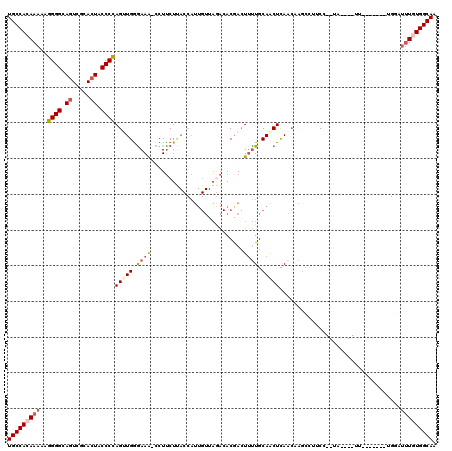
| Location | 6,164,508 – 6,164,611 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -32.68 |
| Energy contribution | -32.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
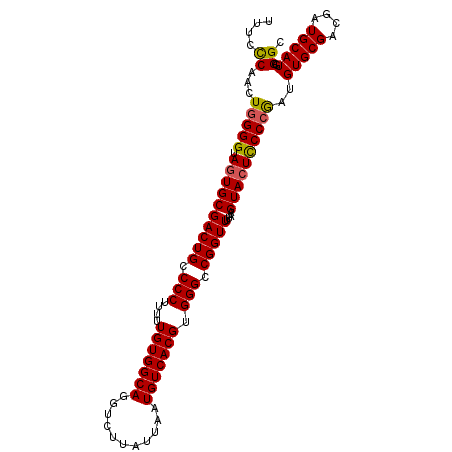
>2L_DroMel_CAF1 6164508 103 + 22407834 UUUUUCAUCUGGGGUAGUGCGACUGCCCCUUUUUGUGGCAGGUCUUAUUAAUGUCACGUGGGCCGGUUUUAAGUACUCCCCGAUGUGCGACGAUGCAUGGGGC .....((((.((((.((((((((((.(((....(((((((...........))))))).))).)))))....)))))))))))))(((......)))...... ( -35.50) >DroSec_CAF1 917 103 + 1 UUUCCCAACUGGGGUAGUGCGACUGCCCCUUUUUGUGGCAGGUCUUAUUAAUGUCACGUGGGCCGGUUUUAAGUACUCCCCGAUGUGCGACGAUGCAUGGGGC ..((((...(((((.((((((((((.(((....(((((((...........))))))).))).)))))....))))))))))..((((......)))))))). ( -39.30) >DroSim_CAF1 10289 103 + 1 UUUCCCAACUGGGGUAGUGCGACUGCCCCUUUUUGUGGCAGGUCUUAUUAAUGUCACGUGGGCCGGUUUUAAGUACUCCCCGAUGUGCGACGAUGCAUGCGGC ....((...(((((.((((((((((.(((....(((((((...........))))))).))).)))))....))))))))))..((((......))))..)). ( -35.30) >DroEre_CAF1 366 103 + 1 UUUCCCAACUGGGGUACUGCGACUGCCCCUUUUUGUGGCAGGUCUUAUUAAUGUCACGUGGGCCGGUUUUAAGUACUCCCCAAUGUGCGACGAUGCAUGCGGC ....((....((((((((..(((((.(((....(((((((...........))))))).))).)))))...)))))))).....((((......))))..)). ( -34.20) >DroYak_CAF1 10393 103 + 1 CUUCCCAUCUAGGGUAGUGCGACUGCCCCUUUUUGUGGCAGGUCUUAUUAAUGUCACGUGGGCCGGUUUUAAGUACUUCCCGAUGUGCGGCGAUGCAUGCGGC .....((((..(((.((((((((((.(((....(((((((...........))))))).))).)))))....))))).))))))).((.(((.....))).)) ( -33.60) >consensus UUUCCCAACUGGGGUAGUGCGACUGCCCCUUUUUGUGGCAGGUCUUAUUAAUGUCACGUGGGCCGGUUUUAAGUACUCCCCGAUGUGCGACGAUGCAUGCGGC ....((...(((((.((((((((((.(((....(((((((...........))))))).))).)))))....))))))))))..(((((....)))))..)). (-32.68 = -32.60 + -0.08)

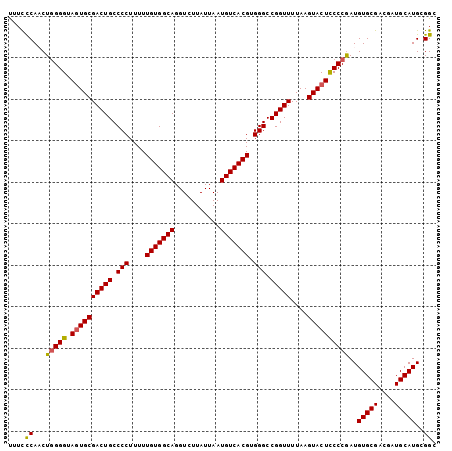
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:42 2006