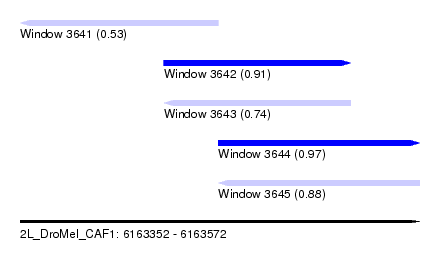
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,163,352 – 6,163,572 |
| Length | 220 |
| Max. P | 0.969136 |

| Location | 6,163,352 – 6,163,461 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.09 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -19.29 |
| Energy contribution | -20.89 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6163352 109 - 22407834 GGCUAGUGCUGCUGGUGCUUGAGUGUG----------UGCGUAGCCCCCAUUAGCCAA-AGGAUUUAUAUUACAAGCGCACACAUAAAAAGCACCGCAGGAUAUUCGGUUUUAUUUGAAU .....((.((((.(((((((..(((((----------(((((............((..-.)).............))))))))))...))))))))))).))((((((......)))))) ( -36.61) >DroPse_CAF1 11938 112 - 1 UCCGAAUGCUGUGUGUG--------UGUGUGGGUGUUUUCGUGGCCCCCAUUAGCCAAAAGGAUUUAUAUUACAAGCGCACACAUAAAAAGCACCGCAGGAUAUUUUGUUUUAUGUGAAU (((...((((.((((((--------(((((.(((((.(((.((((........))))...)))...)))))....)))))))))))...)))).....)))................... ( -33.00) >DroSim_CAF1 9153 109 - 1 GGCGAGUGCUGCUGGUGCUUGAGUGUG----------UGCGUAGCCCCCAUUAGCCAA-AGGAUUUAUAUUACAAGCGCACACAUAAAAAGCACCGCAGGAUAUUCGGUUUUAUUUGAAU ..((((((((((.(((((((..(((((----------(((((............((..-.)).............))))))))))...)))))))))))..))))))............. ( -38.01) >DroYak_CAF1 9186 109 - 1 GGCGAGUGCUGCUGGUGCUUGAGUGUG----------UGCGUAGCCCCCAUUAGCCAA-AGGAUUUAUAUUACAAGCGCACACAUAAAAAGCACCGCAGGAUAUUCGGUUUUAUUUGAAU ..((((((((((.(((((((..(((((----------(((((............((..-.)).............))))))))))...)))))))))))..))))))............. ( -38.01) >DroPer_CAF1 11934 120 - 1 UCCGAAUGCUGUGUGUGUGUGAGUGUGUGUGGGUGUUUUCGUGGCCCCCAUUAGCCAAAAGGAUUUAUAUUACAAGCGCACACAUAAAAAGCACCGCAGGAUAUUUUGUUUUAUGUGAAU (((...((((.((((((((((..(((((((((((.((((..((((........)))))))).))))))).))))..))))))))))...)))).....)))................... ( -38.00) >consensus GGCGAGUGCUGCUGGUGCUUGAGUGUG__________UGCGUAGCCCCCAUUAGCCAA_AGGAUUUAUAUUACAAGCGCACACAUAAAAAGCACCGCAGGAUAUUCGGUUUUAUUUGAAU .....((.((((.(((((((..(((((..........(((((............((....)).............))))))))))...))))))))))).)).................. (-19.29 = -20.89 + 1.60)

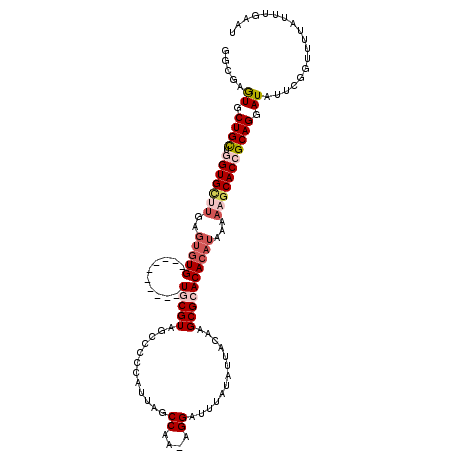
| Location | 6,163,431 – 6,163,534 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -26.69 |
| Energy contribution | -26.69 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6163431 103 + 22407834 GCACACACUCAAGCACCAGCAGCACUAGCCAGAGCUCCAACUGUUCGCAGCAGGAGCUCUCUGCUCCGAGCUCCACUGCAUGCCUCUUGCUCCCAGCUCCC-------GG ((..........)).(((((((........((((((((..(((....)))..)))))))))))))..(((((.....(((.......)))....)))))..-------)) ( -32.10) >DroSim_CAF1 9232 105 + 1 GCACACACUCAAGCACCAGCAGCACUCGCCAGAGCUCCAACUGUUCGCAGCAGGAGCUCUCUGCUCCGAGCUCC-----AUGCCUCUUGCUCCCAGCUCCCAGCAACCAG ............((...(((((........((((((((..(((....)))..)))))))))))))..(((((..-----..((.....))....)))))...))...... ( -31.90) >DroYak_CAF1 9265 105 + 1 GCACACACUCAAGCACCAGCAGCACUCGCCAGAGCUCCCACUGUUCGCAGCAGGAGCUCUCUGCUCCCAGCUCC-----AUGCCUCGUGCUCCGUGCCUUGUGCUCCGAG ((((((((...(((((.(((((........((((((((..(((....)))..)))))))))))))....((...-----..))...)))))..)))...)))))...... ( -37.70) >consensus GCACACACUCAAGCACCAGCAGCACUCGCCAGAGCUCCAACUGUUCGCAGCAGGAGCUCUCUGCUCCGAGCUCC_____AUGCCUCUUGCUCCCAGCUCCC_GC__C_AG (((........(((...(((((........((((((((..(((....)))..)))))))))))))....)))........))).....((.....))............. (-26.69 = -26.69 + -0.00)

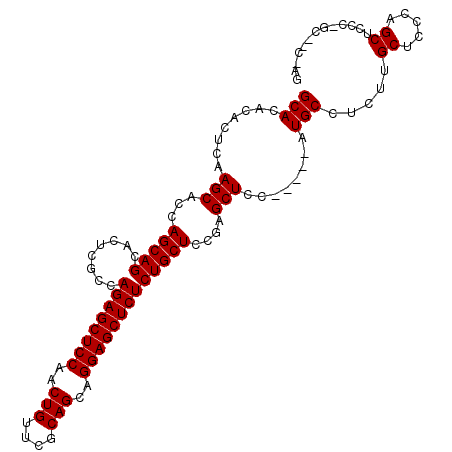
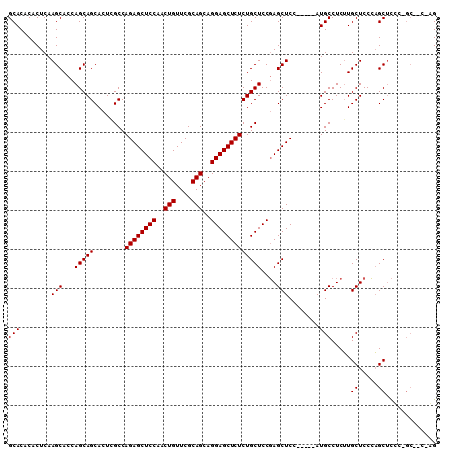
| Location | 6,163,431 – 6,163,534 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -34.90 |
| Energy contribution | -35.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6163431 103 - 22407834 CC-------GGGAGCUGGGAGCAAGAGGCAUGCAGUGGAGCUCGGAGCAGAGAGCUCCUGCUGCGAACAGUUGGAGCUCUGGCUAGUGCUGCUGGUGCUUGAGUGUGUGC .(-------(((.((..(.((((...(((.(((((((((((((........))))))).))))))..(((........))))))..)))).)..)).))))......... ( -43.50) >DroSim_CAF1 9232 105 - 1 CUGGUUGCUGGGAGCUGGGAGCAAGAGGCAU-----GGAGCUCGGAGCAGAGAGCUCCUGCUGCGAACAGUUGGAGCUCUGGCGAGUGCUGCUGGUGCUUGAGUGUGUGC ....(((((..(((((..(.((.....)).)-----..)))))..)))))((((((((.((((....)))).))))))))..((((..(.....)..))))......... ( -40.90) >DroYak_CAF1 9265 105 - 1 CUCGGAGCACAAGGCACGGAGCACGAGGCAU-----GGAGCUGGGAGCAGAGAGCUCCUGCUGCGAACAGUGGGAGCUCUGGCGAGUGCUGCUGGUGCUUGAGUGUGUGC ......((((...((((.((((((.((((((-----...((((....)..((((((((..(((....)))..)))))))))))..))))..)).))))))..)))))))) ( -49.70) >consensus CU_G__GC_GGGAGCUGGGAGCAAGAGGCAU_____GGAGCUCGGAGCAGAGAGCUCCUGCUGCGAACAGUUGGAGCUCUGGCGAGUGCUGCUGGUGCUUGAGUGUGUGC .............((.....(((..((((((.....(.(((.(...((..((((((((..(((....)))..)))))))).))..).))).)..))))))...)))..)) (-34.90 = -35.23 + 0.33)


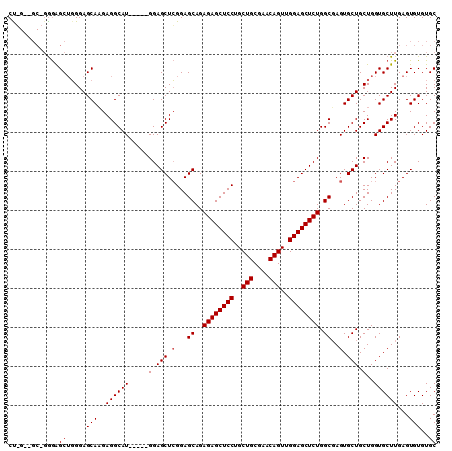
| Location | 6,163,461 – 6,163,572 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -30.08 |
| Energy contribution | -30.75 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
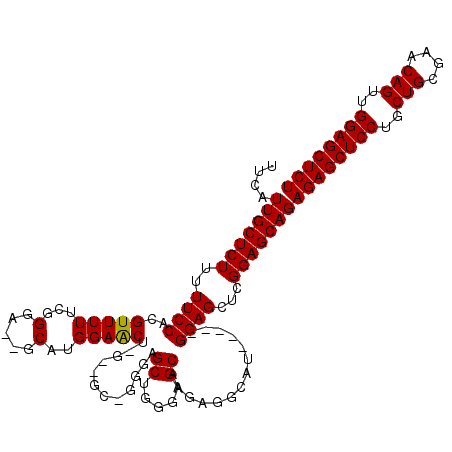
>2L_DroMel_CAF1 6163461 111 + 22407834 AGAGCUCCAACUGUUCGCAGCAGGAGCUCUCUGCUCCGAGCUCCACUGCAUGCCUCUUGCUCCCAGCUCCC-------GGUUCCAUGC--UUCCGAAAAACGUGGAAAAAGAGCAAUGAA ((((((((..(((....)))..)))))))).(((((.(((((.....(((.......)))....)))))..-------..(((((((.--..........)))))))...)))))..... ( -39.30) >DroSim_CAF1 9262 113 + 1 AGAGCUCCAACUGUUCGCAGCAGGAGCUCUCUGCUCCGAGCUCC-----AUGCCUCUUGCUCCCAGCUCCCAGCAACCAGUUCCAUGC--UCCCGAAAAACGUGGAAAAAGAGCAAUGAA ((((((((..(((....)))..)))))))).(((((.(((((..-----..((.....))....)))))...........(((((((.--..........)))))))...)))))..... ( -37.80) >DroYak_CAF1 9295 115 + 1 AGAGCUCCCACUGUUCGCAGCAGGAGCUCUCUGCUCCCAGCUCC-----AUGCCUCGUGCUCCGUGCCUUGUGCUCCGAGCUCCAUGCCAUCCCGAAAAACGGGGAAAAAGAGCAAUGAA ((((((((..(((....)))..)))))))).(((((..(((((.-----..(((..(.((.....)))..).))...)))))........(((((.....))))).....)))))..... ( -40.90) >consensus AGAGCUCCAACUGUUCGCAGCAGGAGCUCUCUGCUCCGAGCUCC_____AUGCCUCUUGCUCCCAGCUCCC_GC__C_AGUUCCAUGC__UCCCGAAAAACGUGGAAAAAGAGCAAUGAA ((((((((..(((....)))..)))))))).(((((.((((.................))))...................((((((.............))))))....)))))..... (-30.08 = -30.75 + 0.67)

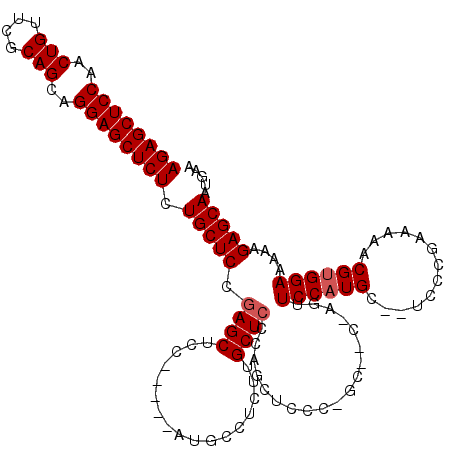
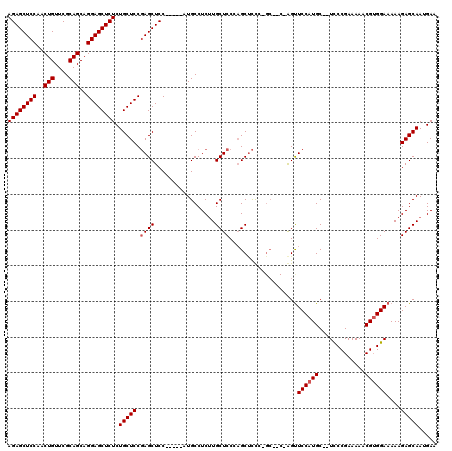
| Location | 6,163,461 – 6,163,572 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -34.55 |
| Energy contribution | -34.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6163461 111 - 22407834 UUCAUUGCUCUUUUUCCACGUUUUUCGGAA--GCAUGGAACC-------GGGAGCUGGGAGCAAGAGGCAUGCAGUGGAGCUCGGAGCAGAGAGCUCCUGCUGCGAACAGUUGGAGCUCU ....((((((...(((((.(((((...)))--)).))))).(-------(((.((((.(.((.....)).).))))....))))))))))((((((((.((((....)))).)))))))) ( -45.10) >DroSim_CAF1 9262 113 - 1 UUCAUUGCUCUUUUUCCACGUUUUUCGGGA--GCAUGGAACUGGUUGCUGGGAGCUGGGAGCAAGAGGCAU-----GGAGCUCGGAGCAGAGAGCUCCUGCUGCGAACAGUUGGAGCUCU ....(((((((..(((((.((((.....))--)).)))))(..(((......)))..)((((.........-----...)))))))))))((((((((.((((....)))).)))))))) ( -43.00) >DroYak_CAF1 9295 115 - 1 UUCAUUGCUCUUUUUCCCCGUUUUUCGGGAUGGCAUGGAGCUCGGAGCACAAGGCACGGAGCACGAGGCAU-----GGAGCUGGGAGCAGAGAGCUCCUGCUGCGAACAGUGGGAGCUCU ....((((((((.(((((((.....))).(((.(.((..((((.(.((.....)).).)))).)).).)))-----))))..))))))))((((((((..(((....)))..)))))))) ( -52.00) >consensus UUCAUUGCUCUUUUUCCACGUUUUUCGGGA__GCAUGGAACU_G__GC_GGGAGCUGGGAGCAAGAGGCAU_____GGAGCUCGGAGCAGAGAGCUCCUGCUGCGAACAGUUGGAGCUCU ....(((((((..((((..(((((...(.....)..)))))............((.....))..............))))...)))))))((((((((..(((....)))..)))))))) (-34.55 = -34.33 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:39 2006