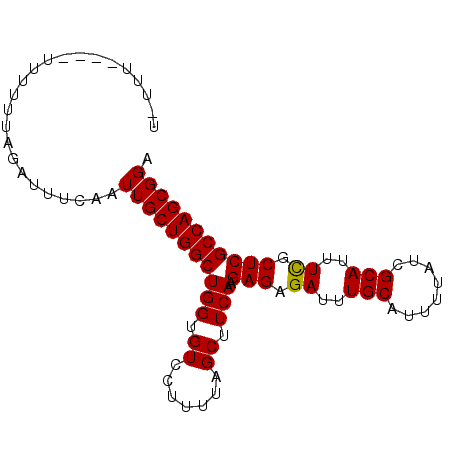
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 635,905 – 635,995 |
| Length | 90 |
| Max. P | 0.999941 |

| Location | 635,905 – 635,995 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.85 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 635905 90 + 22407834 UCCGCUGGCGAGCGAAAUGCGAUAAAAUGCAAAUCUCUCUUGAAGCUAAAAGGACACCAGCCAGCAAUUGAAAUCUAAAACAAAAAAAA-A ...(((((((((.((..((((......))))..)).)))............((...)).))))))........................-. ( -20.00) >DroSec_CAF1 24669 83 + 1 UCCGCUGGCGAGCGAAAUGCGAUAAAAUGCAAAUCUCUCUUGAAGCUAAAAGUACACCAGCCAGCAAUUGAAAUCUAAAAAC--------A ...(((((((((.((..((((......))))..)).))).....((.....))......)))))).................--------. ( -19.60) >DroSim_CAF1 24568 83 + 1 UCCGCUGGCGAGCGAAAUGCGAUAAAAUGCAAAUCUCUCUUGAAGCUAAAAGGACACCAGCCAGCAAUUGAAAUCUAAAAAC--------A ...(((((((((.((..((((......))))..)).)))............((...)).)))))).................--------. ( -20.00) >DroEre_CAF1 29077 91 + 1 UCCGCUGGCGAGCAAAAUGCGAUAAAAUGCAAAUCUCUCUUGAAGCUAAAAGGACACCAGCCAGCAAUUGAAAUCUAAAAAAAAAAAAACA ...((((((.(((....((((......))))..((......)).)))....((...)).)))))).......................... ( -18.00) >DroYak_CAF1 28244 89 + 1 UCCGCUGGCGAGCAAAAUGCGAUAAAAUGCAAAUCUCUCUUGAAGCUAAAAGGACACCAGCCAGCAAUUGAAAUCUAAAAAAC--GAAAUA ...((((((.(((....((((......))))..((......)).)))....((...)).))))))..................--...... ( -18.00) >consensus UCCGCUGGCGAGCGAAAUGCGAUAAAAUGCAAAUCUCUCUUGAAGCUAAAAGGACACCAGCCAGCAAUUGAAAUCUAAAAAA____AAA_A ...(((((((((.((..((((......))))..)).)))............(.....).)))))).......................... (-17.52 = -17.92 + 0.40)

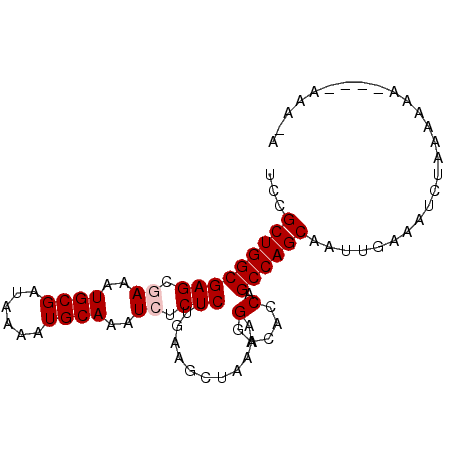
| Location | 635,905 – 635,995 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.85 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 635905 90 - 22407834 U-UUUUUUUUGUUUUAGAUUUCAAUUGCUGGCUGGUGUCCUUUUAGCUUCAAGAGAGAUUUGCAUUUUAUCGCAUUUCGCUCGCCAGCGGA .-......................(((((((((((.((.......)).))).(((.((..(((........)))..)).))))))))))). ( -23.10) >DroSec_CAF1 24669 83 - 1 U--------GUUUUUAGAUUUCAAUUGCUGGCUGGUGUACUUUUAGCUUCAAGAGAGAUUUGCAUUUUAUCGCAUUUCGCUCGCCAGCGGA .--------...............(((((((((((.((.......)).))).(((.((..(((........)))..)).))))))))))). ( -23.40) >DroSim_CAF1 24568 83 - 1 U--------GUUUUUAGAUUUCAAUUGCUGGCUGGUGUCCUUUUAGCUUCAAGAGAGAUUUGCAUUUUAUCGCAUUUCGCUCGCCAGCGGA .--------...............(((((((((((.((.......)).))).(((.((..(((........)))..)).))))))))))). ( -23.10) >DroEre_CAF1 29077 91 - 1 UGUUUUUUUUUUUUUAGAUUUCAAUUGCUGGCUGGUGUCCUUUUAGCUUCAAGAGAGAUUUGCAUUUUAUCGCAUUUUGCUCGCCAGCGGA ........................(((((((((((.((.......)).))).(((..(..(((........)))..)..))))))))))). ( -20.90) >DroYak_CAF1 28244 89 - 1 UAUUUC--GUUUUUUAGAUUUCAAUUGCUGGCUGGUGUCCUUUUAGCUUCAAGAGAGAUUUGCAUUUUAUCGCAUUUUGCUCGCCAGCGGA ......--................(((((((((((.((.......)).))).(((..(..(((........)))..)..))))))))))). ( -20.90) >consensus U_UUU____UUUUUUAGAUUUCAAUUGCUGGCUGGUGUCCUUUUAGCUUCAAGAGAGAUUUGCAUUUUAUCGCAUUUCGCUCGCCAGCGGA ........................(((((((((((.((.......)).))).(((.((..(((........)))..)).))))))))))). (-22.48 = -22.24 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:33 2006