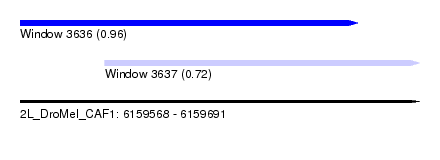
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,159,568 – 6,159,691 |
| Length | 123 |
| Max. P | 0.955426 |

| Location | 6,159,568 – 6,159,672 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.79 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6159568 104 + 22407834 UU----------GGCAGCUACC--CUGCUGUCAGA--GCCAUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC-UAAAAGUCAAAGACA-AA ((----------((((((....--..)))))))).--.......(((((......)))))..((((((((((....................)))))))))-)....(((...))).-.. ( -22.65) >DroPse_CAF1 6937 120 + 1 GUCCACGCAGUCGCUCGCUUCCUUCGGCUGUCAAAAGGACGUCACAAAAAUGACAUUUUCCUGGCUAAUUACAAUUUCAAAUUAAAUCACGAGUAAUUAGCCCGAAAGUCAAAGCCAAAA ((((..(((((((...........))))))).....))))((((......))))((((((..((((((((((....................)))))))))).))))))........... ( -30.25) >DroSim_CAF1 5401 104 + 1 UU----------GGCAGCUACC--CUGCUGUCAGA--GCCAUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC-UGAAAGUCAAAGACA-AA ((----------((((((....--..)))))))).--.......(((((......))))).(((((((((((....................)))))))))-))...(((...))).-.. ( -24.95) >DroYak_CAF1 5358 104 + 1 UU----------GGCAGCUACC--CGGCUGUCAGA--GCCAUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC-UGAAAGUCAAAGACA-AA ((----------(((((((...--.))))))))).--.......(((((......))))).(((((((((((....................)))))))))-))...(((...))).-.. ( -25.15) >DroPer_CAF1 7001 120 + 1 GUCCACGCUGUCGCUCGCUUCCUUCGGCUGUCAAAAGGACGUCACAAAAAUGACAUUUUCCCGGCUAAUUACAAUUUCAAAUUAAAUCACGAGUAAUUAGCCCGAAAGUCAAAGCCAAAA ((((..((....))..(((......)))........))))((((......))))((((((..((((((((((....................)))))))))).))))))........... ( -27.05) >consensus UU__________GGCAGCUACC__CGGCUGUCAGA__GCCAUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC_UGAAAGUCAAAGACA_AA ............((((((........)))))).............................(((((((((((....................))))))))).))................ (-14.39 = -14.79 + 0.40)



| Location | 6,159,594 – 6,159,691 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -10.33 |
| Energy contribution | -10.37 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6159594 97 + 22407834 AUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC-UAAAAGUCAAAGACA-AACAUGCCAAACA--------------------AUGGGGC-G ...................(((((((((((((....................)))))))))-)....(((...))).-......(((....--------------------.))))))-. ( -17.55) >DroPse_CAF1 6977 119 + 1 GUCACAAAAAUGACAUUUUCCUGGCUAAUUACAAUUUCAAAUUAAAUCACGAGUAAUUAGCCCGAAAGUCAAAGCCAAAACAUGGCAAACACACGACAGGGAUGGUAUGGGAUGGGGC-A ((((......))))((((((..((((((((((....................)))))))))).))))))....(((....((((.((..(.(......))..)).))))......)))-. ( -27.85) >DroSim_CAF1 5427 97 + 1 AUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC-UGAAAGUCAAAGACA-AACAUGGCAAACA--------------------AUGGGGC-G .............(((((((((((((((((((....................)))))))))-)....(((...))).-.....)))))..)--------------------)))....-. ( -19.75) >DroYak_CAF1 5384 98 + 1 AUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC-UGAAAGUCAAAGACA-AACAUGGCAAACA--------------------AUGGGGCCC .............(((((((((((((((((((....................)))))))))-)....(((...))).-.....)))))..)--------------------)))...... ( -19.75) >DroPer_CAF1 7041 114 + 1 GUCACAAAAAUGACAUUUUCCCGGCUAAUUACAAUUUCAAAUUAAAUCACGAGUAAUUAGCCCGAAAGUCAAAGCCAAAACCUGGCAAACACACGACACGGCU-----GGGAUGGGGC-A ((((......))))((((((..((((((((((....................)))))))))).))))))....(((..(.((..((..............)).-----.)).)..)))-. ( -30.39) >consensus AUCACAAAAAUUACAUUUUGCCGGCUAAUUACAAUUUCAAAUUAAAUCACAAGUAAUUAGC_UGAAAGUCAAAGACA_AACAUGGCAAACA____________________AUGGGGC_A ....................((((((((((((....................)))))))))......((((...........))))..........................)))..... (-10.33 = -10.37 + 0.04)

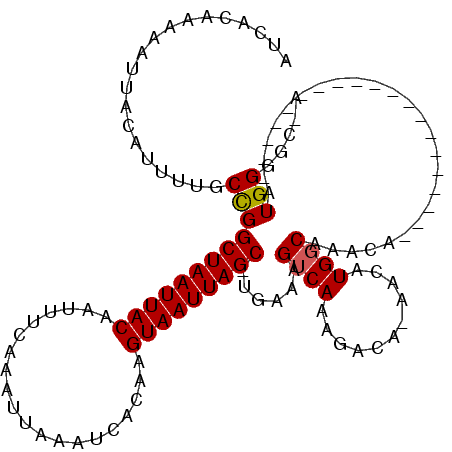

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:32 2006