
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,156,106 – 6,156,257 |
| Length | 151 |
| Max. P | 0.995639 |

| Location | 6,156,106 – 6,156,207 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -4.52 |
| Energy contribution | -5.48 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.18 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6156106 101 - 22407834 AAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGGGUA--AGCCA---------------UGUAUAUAAACUUCCCAAUGGCGCCCGAACAGGCUUAAGACCUU--CGCACAGCU .(((((((..((((....))))...)))))))...(((((((.--.((((---------------(................)))))(((......))).....)))))--))....... ( -27.09) >DroPse_CAF1 2174 118 - 1 AAUUCAAUUAUUGCCCCCGAACAAUAUUGAAUCAGCAGCAGCAGCAGCAACACCAGUCUGGGAGAGAGAGAUGAGAGCUCUAGAGCCAGAGGA--AAACUUAAGACCUUCUCGCACAGCU .(((((((.((((........))))))))))).(((.((....)).((....((..(((((.....((((.(....)))))....))))))).--........((.....))))...))) ( -22.90) >DroSim_CAF1 1966 101 - 1 AAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGGGUA--AGCCA---------------UGUAUAUAAACUUCCCAAUGGCGCCCGAACAGGCUUAAGACCUU--CGCACAGCU .(((((((..((((....))))...)))))))...(((((((.--.((((---------------(................)))))(((......))).....)))))--))....... ( -27.09) >DroYak_CAF1 1935 101 - 1 AAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGGGUA--AGCCA---------------UGUAUAUAAACUUCCCAGUGGCGCCCGAACAGGCUUAAGACCUU--CGCACAGCU .(((((((..((((....))))...)))))))...(((((((.--.((((---------------(................)))))(((......))).....)))))--))....... ( -26.79) >DroPer_CAF1 2155 118 - 1 AAUUCAAUUAUUGCCCCCGAACAAUAUUGAAUCAGCAGCAGCAGCAGCAACACCAGUCUGGGAGAGAGAGAUGAGAGCUCUAGAGGCAGAGGA--AAACUUAAGACCUUCUCGCACAGCU .(((((((.((((........))))))))))).(((.((....)).((....((.....))((((.((((.(....)))))..((((..(((.--...)))..).)))))))))...))) ( -23.60) >consensus AAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGGGUA__AGCCA_______________UGUAUAUAAACUUCCCAGUGGCGCCCGAACAGGCUUAAGACCUU__CGCACAGCU .(((((((..((((....))))...))))))).......(((....)))....................................................................... ( -4.52 = -5.48 + 0.96)



| Location | 6,156,167 – 6,156,257 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.02 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -3.72 |
| Energy contribution | -4.52 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.16 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6156167 90 - 22407834 ACCGCCUCUU-CACAACCACAUUUCCAA-----------------------------AGGAAUGUCCCAGUCAUCUAUUAAAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGG ....((((..-.......(((((((...-----------------------------.)))))))................(((((((..((((....))))...)))))))....)))) ( -16.50) >DroPse_CAF1 2252 120 - 1 UUCUGCUCUUCCAUAAUUCCCUCUCCAAAGCAGCUGCCAGCGCGGUUUCCUUUAAAGCCGCGUUGCCCAGCCAGCUAUUAAAUUCAAUUAUUGCCCCCGAACAAUAUUGAAUCAGCAGCA ..(((((.....................(((.((((.(((((((((((......)))))))))))..))))..))).....(((((((.((((........))))))))))).))))).. ( -35.50) >DroSim_CAF1 2027 90 - 1 ACCACCUCUU-CACAACCACAUUUCCAA-----------------------------CGGAAUGUCCCAGCCAUCUAUUAAAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGG ....((((..-.......(((((((...-----------------------------.)))))))................(((((((..((((....))))...)))))))....)))) ( -16.70) >DroYak_CAF1 1996 90 - 1 ACCACCACUU-CACUACCAUAUUUCCAA-----------------------------AGGAAUUUCCCAGCCAUCUAUUAAAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGG .......(((-(................-----------------------------.((......)).............(((((((..((((....))))...)))))))....)))) ( -11.50) >DroPer_CAF1 2233 120 - 1 UUCUGCUCUUCCAUAAAUCCCUCUCCAAAGCAGCUGCCAGCGCGGUUUCCUUUAAAGCCGCGUUGCCCAGCCAGCUAUUAAAUUCAAUUAUUGCCCCCGAACAAUAUUGAAUCAGCAGCA ..(((((.....................(((.((((.(((((((((((......)))))))))))..))))..))).....(((((((.((((........))))))))))).))))).. ( -35.50) >consensus ACCACCUCUU_CACAACCACAUUUCCAA_____________________________CGGAAUUUCCCAGCCAUCUAUUAAAUUCAAUUAUUGCCCAAGCAAAAUAUUGAAUCAACGAGG .................................................................................(((((((..((((....))))...)))))))........ ( -3.72 = -4.52 + 0.80)

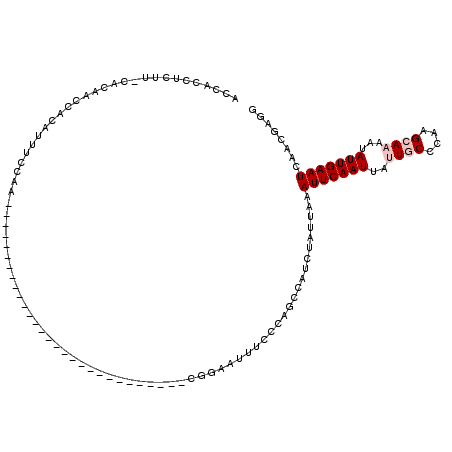
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:27 2006