
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,155,942 – 6,156,066 |
| Length | 124 |
| Max. P | 0.996654 |

| Location | 6,155,942 – 6,156,037 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -39.86 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6155942 95 + 22407834 ----CGGGG-------------------CUUGUGGGG-GUUGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG-AAAACCCAGCUGCCUCCAAGGAGCAACAAAA ----.(..(-------------------(((.(((((-((.((((.(((.((((((((.((..((((((....)))))))))))))))-).)))..)))))))))))..))))..).... ( -37.10) >DroPse_CAF1 1989 118 + 1 GGGCUGGGGCUGCGGGGUCAGGAUUGGACUU-CUGGG-GGUGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUGGAAAACCCAGCUGCCUUAAAGGAGCAGCCAGC ..(((((.(((.(((((((.......)))))-)..((-((..(((.(((.((((((((.((..((((((....))))))))))))))))..)))..)))..))))...).)))..))))) ( -43.70) >DroSim_CAF1 1802 95 + 1 ----CGGGG-------------------CUUGUGGGG-GGUGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG-AAAACCCAGCUGCCUCCAAGGAGCAACAAAA ----.(..(-------------------(((.(.(((-((..(((.(((.((((((((.((..((((((....)))))))))))))))-).)))..)))..))))).).))))..).... ( -40.50) >DroYak_CAF1 1760 95 + 1 ----U-UGG-------------------CUUGCGGGGGGUUGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG-AAAACCCAGCUGCCUCCAAGGAGCAACAAAA ----.-..(-------------------(((...((((((.((((.(((.((((((((.((..((((((....)))))))))))))))-).)))..))))))))))...))))....... ( -33.80) >DroPer_CAF1 1973 115 + 1 ----UGGGGCUGCGGGGUCAGGAUUGGACUUCCUGGG-GGUGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUGGAAAACCCAGCUGCCUUAAAGGAGCAGCCAGC ----...((((((..((((.......))))((((..(-((..(((.(((.((((((((.((..((((((....))))))))))))))))..)))..)))..)))...))))))))))... ( -44.20) >consensus ____UGGGG___________________CUUGCGGGG_GGUGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG_AAAACCCAGCUGCCUCCAAGGAGCAACAAAA ............................(((...(((.(((((((.(((..(((((((.((..((((((....)))))))))))))))...)))..))))))))))...)))........ (-21.60 = -22.16 + 0.56)



| Location | 6,155,958 – 6,156,066 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.18 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -19.10 |
| Energy contribution | -20.35 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6155958 108 + 22407834 UGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG-AAAACCCAGCUGCCUCCAAGGAGCAACAAAAAGAACAGAACGACGUCGGACUGAGGGAAA ..........((((((((.((..((((((....)))))))))))))))-)...(((.(((.((.....)))))...........(((..((....))..))).)))... ( -29.00) >DroPse_CAF1 2027 107 + 1 UGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUGGAAAACCCAGCUGCCUUAAAGGAGCAGCCAGCGGCAGCCAGCAGCAGCCGA--AAAGGAAA ((((.(((((......)))))...)))).....(((((.(((....(((....(((.(((((((....)).)))))..).))...)))...))))))))--........ ( -33.40) >DroSim_CAF1 1818 108 + 1 UGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG-AAAACCCAGCUGCCUCCAAGGAGCAACAAAAAGAACAGAACGUCGUCGGACUGAGGGAAA ..........((((((((.((..((((((....)))))))))))))))-)...(((.(((.((.....)))))...........(((..((....))..))).)))... ( -28.40) >DroPer_CAF1 2008 107 + 1 UGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUGGAAAACCCAGCUGCCUUAAAGGAGCAGCCAGCGGCAGCCAGCAGCAACCGA--AAAGGAAA ((((.(((((......)))))...)))).....(((((.(((....(((.....)))(((((((....)).))))).....))))))))......((..--...))... ( -31.30) >consensus UGGCUUGUUAUUAUUUUGACACUUGCCAAACAAUUGGCAUGCGAAAUG_AAAACCCAGCUGCCUCAAAGGAGCAACAAAAAGAACACAACAGCAUCCGA__AAAGGAAA ..(((((((..(((((((.((..((((((....))))))))))))))).........(((.((.....))))))))))))............................. (-19.10 = -20.35 + 1.25)

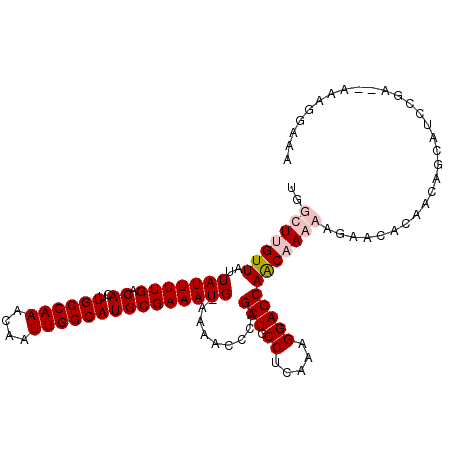
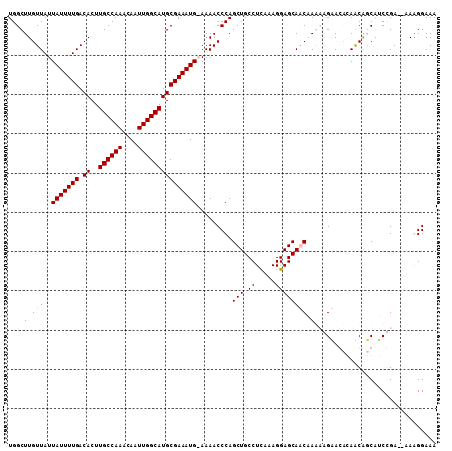
| Location | 6,155,958 – 6,156,066 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.18 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -22.27 |
| Energy contribution | -21.52 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6155958 108 - 22407834 UUUCCCUCAGUCCGACGUCGUUCUGUUCUUUUUGUUGCUCCUUGGAGGCAGCUGGGUUUU-CAUUUCGCAUGCCAAUUGUUUGGCAAGUGUCAAAAUAAUAACAAGCCA ...(((.(((..((....))..)))........(((((((....)).))))).)))....-.....(((.((((((....)))))).)))................... ( -27.00) >DroPse_CAF1 2027 107 - 1 UUUCCUUU--UCGGCUGCUGCUGGCUGCCGCUGGCUGCUCCUUUAAGGCAGCUGGGUUUUCCAUUUCGCAUGCCAAUUGUUUGGCAAGUGUCAAAAUAAUAACAAGCCA ........--..((((....(..((((((...((.....)).....))))))..)(((........(((.((((((....)))))).)))..........))).)))). ( -34.87) >DroSim_CAF1 1818 108 - 1 UUUCCCUCAGUCCGACGACGUUCUGUUCUUUUUGUUGCUCCUUGGAGGCAGCUGGGUUUU-CAUUUCGCAUGCCAAUUGUUUGGCAAGUGUCAAAAUAAUAACAAGCCA ...(((.(((..((....))..)))........(((((((....)).))))).)))....-.....(((.((((((....)))))).)))................... ( -26.00) >DroPer_CAF1 2008 107 - 1 UUUCCUUU--UCGGUUGCUGCUGGCUGCCGCUGGCUGCUCCUUUAAGGCAGCUGGGUUUUCCAUUUCGCAUGCCAAUUGUUUGGCAAGUGUCAAAAUAAUAACAAGCCA ........--..((((....(..((((((...((.....)).....))))))..)(((........(((.((((((....)))))).)))..........))).)))). ( -32.47) >consensus UUUCCCUC__UCCGACGCCGCUCGCUGCCGCUGGCUGCUCCUUGAAGGCAGCUGGGUUUU_CAUUUCGCAUGCCAAUUGUUUGGCAAGUGUCAAAAUAAUAACAAGCCA ...................(((........(..((((((.......))))))..)(((........(((.((((((....)))))).)))..........))).))).. (-22.27 = -21.52 + -0.75)

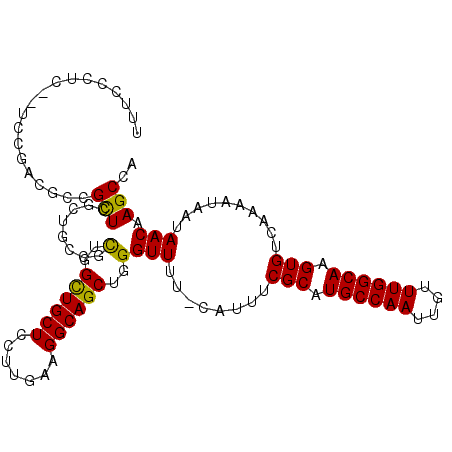
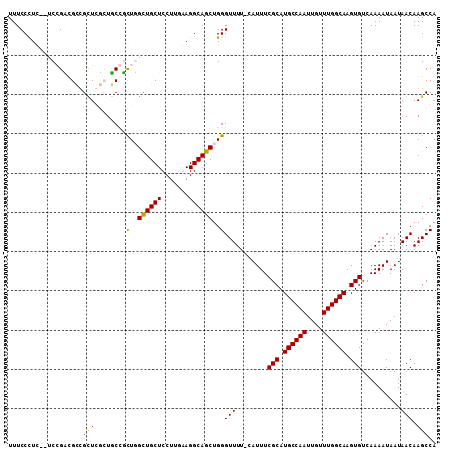
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:25 2006