
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,140,090 – 6,140,250 |
| Length | 160 |
| Max. P | 0.765702 |

| Location | 6,140,090 – 6,140,210 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -56.70 |
| Consensus MFE | -50.24 |
| Energy contribution | -50.55 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
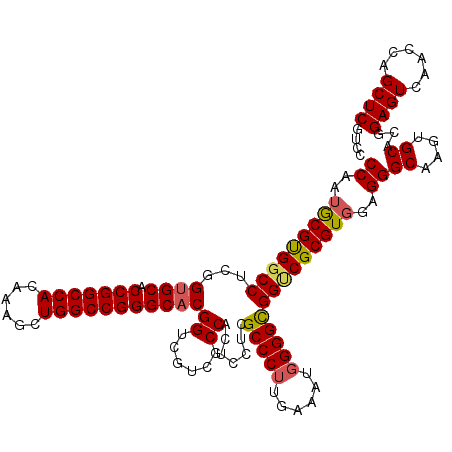
>2L_DroMel_CAF1 6140090 120 - 22407834 CCAAUGCGUGGCCUUGGUGCACCGGCCACAAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUGAAAUGGGGCGGCCGCGUGGAGGGCAAGUGCAUGGAGUCAACCAGCUCGUCC ((...((((((((...((((.(((((((......)))))))))))(((....))).......(((((......))))))))))))))).((((....((.(((......)))))..)))) ( -57.30) >DroSec_CAF1 8902 120 - 1 CCAAUGCGUGGCCUUGGUGCACCGGCCACGAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUGAAAUGGGGCGGUCGCGUUGAGGGCAAGUGCAUGGAGUCAACUAGCUCGUCC .(((((((((((..(.((((.(((((((......))))))))))).).....)))))...(((((((......)))))))..)))))).((((.(((...........))).)))).... ( -56.10) >DroEre_CAF1 10899 120 - 1 CCAAUGCGUGGCCUCGGAGCUCCGGCCACGAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUAAAAUUGGGUGGACGCGUGGAGGGCGUGUGCACGGAGUCAACCAGCUCCUCC ...(((((((((..(((.((.(((((((......)))))))))....)))..)))))...((((((........))))))..))))(((((((((....))((......)).)).))))) ( -54.10) >DroYak_CAF1 9489 120 - 1 CCAAUACGCGGCCUCGGUGCUCCGGCCACAAAGCUGGCCGGGCACGGACGUCGCCACUCCUCGCCCUUGAAAUGGGGCGGUCGCGUGGAGGGCGUGUGCACGGAGUCAACCAGCUCGUCC ....((((((((.((.((((.(((((((......))))))))))).)).)))(((.(((((((((((......)))))))......))))))))))))....((((......)))).... ( -59.30) >consensus CCAAUGCGUGGCCUCGGUGCACCGGCCACAAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUGAAAUGGGGCGGUCGCGUGGAGGGCAAGUGCACGGAGUCAACCAGCUCGUCC ((..(((((((((...((((.(((((((......)))))))))))((......)).......(((((......))))))))))))))..))((....))...((((......)))).... (-50.24 = -50.55 + 0.31)

| Location | 6,140,130 – 6,140,250 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -55.57 |
| Consensus MFE | -51.92 |
| Energy contribution | -51.42 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6140130 120 - 22407834 CGUCAGUUUGGCCAGCUGCUCGUAGUUCUUGGGACAUCGGCCAAUGCGUGGCCUUGGUGCACCGGCCACAAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUGAAAUGGGGCGG .......((((((.(.((((((.......)))).)).))))))).((((((((...((((.(((((((......)))))))))))))))).))).......((((((......)))))). ( -54.60) >DroSec_CAF1 8942 120 - 1 CGUCAGUUUGGCCAGCUGCUCGUAGUUCUUGGGACAUCGGCCAAUGCGUGGCCUUGGUGCACCGGCCACGAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUGAAAUGGGGCGG .....((((((((((((..((((.((...(((..(((((((((.....)))))..))))..))))).))))))))))))))))..(((....)))......((((((......)))))). ( -55.00) >DroEre_CAF1 10939 120 - 1 CGUCAGCUUGGCCAGCUGCUCGUAGUUCUUGGGACACCGGCCAAUGCGUGGCCUCGGAGCUCCGGCCACGAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUAAAAUUGGGUGG .....(((((((((((((((.((.(((.....))))).))).....(((((((..((....))))))))).))))))))))))..(((....)))......(((((........))))). ( -55.80) >DroYak_CAF1 9529 120 - 1 CGUCAGUUUGGCCAGCUGCUCGUAGUCCUUGGGACACUUGCCAAUACGCGGCCUCGGUGCUCCGGCCACAAAGCUGGCCGGGCACGGACGUCGCCACUCCUCGCCCUUGAAAUGGGGCGG .....(((((((.((.((((((.......)))).)))).))))).))(((((.((.((((.(((((((......))))))))))).)).))))).......((((((......)))))). ( -56.90) >consensus CGUCAGUUUGGCCAGCUGCUCGUAGUUCUUGGGACACCGGCCAAUGCGUGGCCUCGGUGCACCGGCCACAAAGCUGGCCGGGCACGGUCGUCGCCACUCCUCGCCCUUGAAAUGGGGCGG .....((((((((.(.((((((.......)))).))).)))))).))(((((..(.((((.(((((((......))))))))))).)..))))).......((((((......)))))). (-51.92 = -51.42 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:21 2006