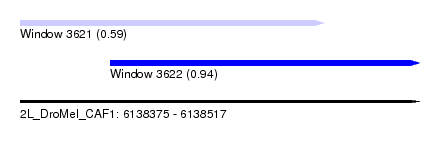
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,138,375 – 6,138,517 |
| Length | 142 |
| Max. P | 0.939819 |

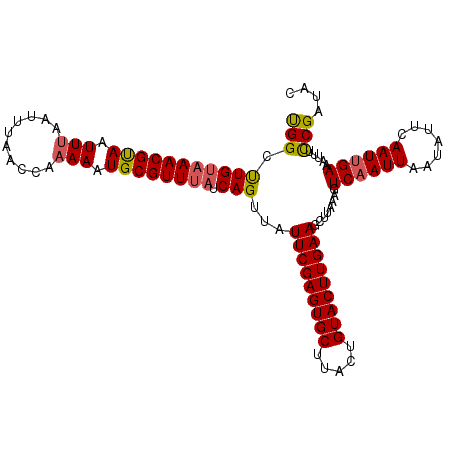
| Location | 6,138,375 – 6,138,483 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.96 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -20.10 |
| Energy contribution | -21.74 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6138375 108 + 22407834 GCAUAUUUGUGUUUGAAUGCACGCG--------UGUGUGUUGGAUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUA ((((((.(((((......))))).)--------)))))....(((((((((((((.(((..........))).)))))))).))))).(((((((((.....)))))))))..... ( -31.00) >DroSec_CAF1 7182 112 + 1 GCAUAUUUGUGUUUGAAUGCACAUGUGUG----UGUGUGUUGGGUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUA ((((((.(((((......))))).)))))----)......(((((((((((((((.(((..........))).))))))))).......((((((((.....)))))))))))))) ( -29.80) >DroSim_CAF1 7371 116 + 1 GCAUAUUUGUGUUUGAAUGCACAUGUGUGUGUGUGUGUGUUGGGUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUA ((((((.(((((......))))).))))))((((.((.(((((((((...........)))))))))))...)))).......(((..(((((((((.....)))))))))))).. ( -30.00) >DroEre_CAF1 8986 106 + 1 --GUAUUUGUGUCUGAGUGCAC----GUG----UGUGUGUCGGCCUGAAAACGCAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUGUUCGAGUGCUUACUGUACUUGAAGCUUA --..........((((.(((((----...----.))))))))).(((((((((((.(((..........))).))))))).))))...(((((((((.....)))))))))..... ( -27.30) >DroYak_CAF1 7754 112 + 1 GCAUAUUUGUGUUUGAAUGCACUUGUGUG----UGUGUGUCGGCUUGUUAACGUAAUUAAAUUUAACCAAAAAUGCGUUUAUCAGUUGUUCGAGUGCUUACUGUACUUGAAGCUUA ((((((..((((......))))..)))))----)......(((((.((.((((((..................)))))).)).)))))(((((((((.....)))))))))..... ( -28.47) >consensus GCAUAUUUGUGUUUGAAUGCACAUGUGUG____UGUGUGUUGGCUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUA ((((.((((.((.((((((((((((........)))))))(((((((......)))))))))))))))))).)))).......(((..(((((((((.....)))))))))))).. (-20.10 = -21.74 + 1.64)

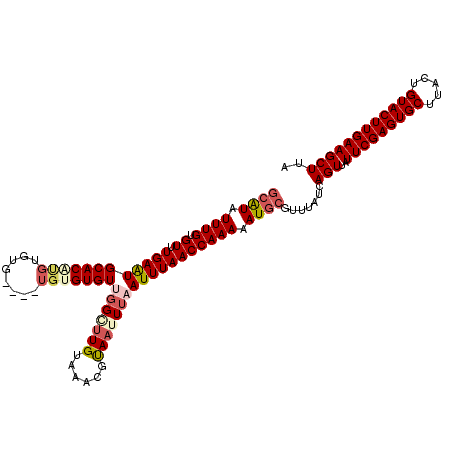
| Location | 6,138,407 – 6,138,517 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -22.41 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6138407 110 + 22407834 UGGAUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUAAAGUCAAUUAAUAUUCAAUGGAAAUUUCCGAUAC ..(((((((((((((.(((..........))).)))))))).))))).(((((((((.....))))))))).......................((((....)))).... ( -22.90) >DroSec_CAF1 7218 110 + 1 UGGGUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUAAAGUCAAUUAAUAUUCAAUUGAAAUUUUCGAUAC (((((((((((((((.(((..........))).))))))))).......((((((((.....))))))))))))))...((((((.......))))))............ ( -21.50) >DroSim_CAF1 7411 110 + 1 UGGGUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUAAAGUCAAUUAAUAUUCAAUUGAAAUUUCCGAUAC .((((((((((((((.(((..........))).)))))))).)))...(((((((((.....)))))))))........((((((.......))))))....)))..... ( -21.60) >DroEre_CAF1 9016 110 + 1 CGGCCUGAAAACGCAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUGUUCGAGUGCUUACUGUACUUGAAGCUUAAAGUCAAUUAAUAUUCAAUUGAAAUAUCCGAUAC (((.(((((((((((.(((..........))).))))))).))))...(((((((((.....)))))))))........((((((.......)))))).....))).... ( -25.90) >DroYak_CAF1 7790 110 + 1 CGGCUUGUUAACGUAAUUAAAUUUAACCAAAAAUGCGUUUAUCAGUUGUUCGAGUGCUUACUGUACUUGAAGCUUAAAGUCAAUUAAUAUUCAAUUGAAAUUUCCGAUAC (((((.((.((((((..................)))))).)).)))))(((((((((.....)))))))))........((((((.......))))))............ ( -20.17) >consensus UGGCUUGUAAACGUAAUUUAAUUUAACCAAAAAUGCGUUUAUCAGUUAUUCGAGUGCUUACUGUACUUGAAGCUUAAAGUCAAUUAAUAUUCAAUUGAAAUUUCCGAUAC (((.(((((((((((.(((..........))).)))))))).)))...(((((((((.....)))))))))........((((((.......)))))).....))).... (-19.66 = -19.74 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:18 2006